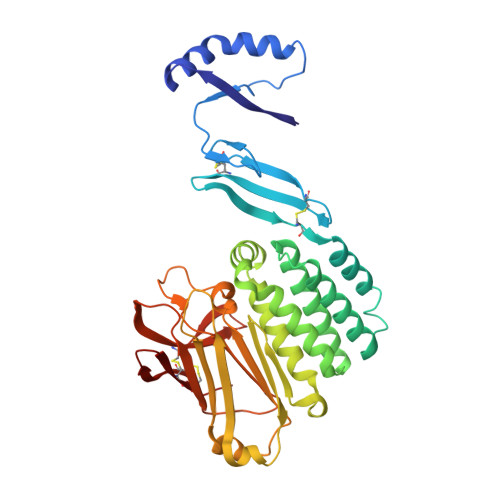
Crystal Structure of the Extracellular Domains of GPR110.
Wang, F., Wang, Y., Qiu, W., Zhang, Q., Yang, H., Song, G.(2023) J Mol Biology 435: 167979-167979
- PubMed: 36716818
- DOI: https://doi.org/10.1016/j.jmb.2023.167979
- Primary Citation of Related Structures:
8HC0 - PubMed Abstract:
Adhesion G protein-coupled receptors (aGPCRs) play a pivotal role in human immune responses, cellular communication, organ development, and other processes. GPR110 belongs to the aGPCR subfamily VI and was initially identified as an oncogene involved in lung and prostate cancers. GPR110 contains tandem adhesion domains at the extracellular region that mediate inter-cellular signaling. However, the structural organization and signaling mechanism for these tandem domains remain unclear. Here, we report the crystal structure of a GPR110 fragment composing the SEA, HormR, and GAIN domains at 2.9 Å resolution. The structure together with MD simulations reveal rigid connections between these domains that are stabilized by complementary interfaces. Strikingly, we found N-linked carbohydrates attached to N389 of the GAIN domain form extensive contacts with the preceding HormR domain. These interactions appear to be critical for folding, as removal of the glycosylation site greatly decreases expression of the GPR110 extracellular fragment. We further demonstrate that the ligand synaptamide fits well within the hydrophobic pocket occupied by the Stachel peptide in the rest state. This suggests that the agonist may function by removing the Stachel peptide which in turn redocks to the orthosteric pocket for receptor activation. Taken together, our structural findings and analyses provide novel insights into the activation mechanism for aGPCRs.
- Shanghai Key Laboratory of Regulatory Biology, Institute of Biomedical Sciences and School of Life Sciences, East China Normal University, Shanghai 200241, China.
Organizational Affiliation: