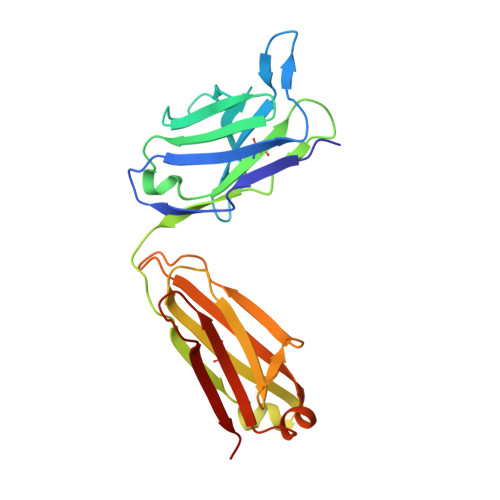
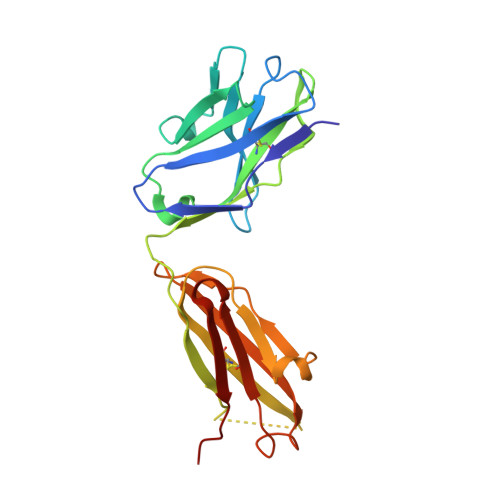
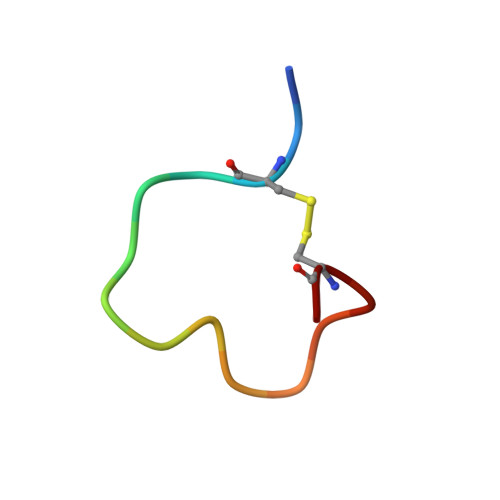
Identification of the binding site and immunoreactivity of anti-A beta antibody 11A1: Comparison with the toxic conformation-specific TxCo-1 antibody.
Fukui, R., Hafizal, U., Kageyama, Y., Irie, Y., Matsushima, Y., Hosoi, K., Nakayama, T., Kaneda, D., Hashizume, Y., Miki, K., Kita, A., Mukaisho, K.I., Kushima, R., Tooyama, I., Irie, K.(2025) Biochem Biophys Res Commun 758: 151655-151655
- PubMed: 40120343
- DOI: https://doi.org/10.1016/j.bbrc.2025.151655
- Primary Citation of Related Structures:
8H8Q - PubMed Abstract:
Since the advent of anti-amyloid β (Aβ) immunotherapy, exemplified by lecanemab, the development of effective therapeutic agents with minimal side effects has become an urgent priority. Over the past two decades, a number of antibodies have been developed that target toxic Aβ species. The 11A1 antibody is one such example, and is made from E22P-Aβ9-35, which is prone to adopt a toxic conformation with a turn at positions 22/23, as an antigen. This antibody is unique in that it stains not only extracellular but also intracellular Aβ in human AD brains. To identify its recognition domain, we performed X-ray crystallography of 11A1 in complex with E22P-Aβ10-34. We found that 11A1 is a novel N-terminal antibody that recognizes Tyr10-His14 of Aβ. Immunohistochemical studies showed that 11A1 stains senile plaques and vascular Aβ aggregates in brain samples of AD patients. On the other hand, 11A1 recognized Aβ aggregates in neurons, astrocytes, perivascular tissue, and microvesicles of non-AD patients, suggesting that 11A1 can detect a wide range of Aβ types regardless of AD pathology. In contrast, the recently developed TxCo-1 antibody, which specifically recognizes the toxic turn at positions 22/23 of Aβ42, stained only senile plaques and vascular Aβ aggregates from AD patients, but not Aβ species from non-AD patients. These results suggest that the toxic turn structure may be one of the key epitopes for achieving high affinity for pathological Aβ aggregates while minimizing nonspecific binding to aggregates unrelated to pathology.
- Department of Pathology (Human Pathology), Shiga University of Medical Science, Otsu, 520-2192, Japan.
Organizational Affiliation: