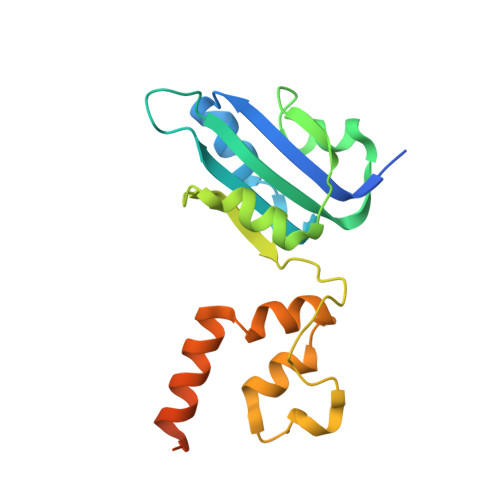
Crystal structure of the dimerized of porcine circovirus type II replication-related protein Rep'.
Guan, S., Tian, A., Jing, H., Yuan, H., Jia, H., Shi, Y., Chen, H., Cao, S., Peng, G., Song, Y.(2023) Proteins 91: 1130-1139
- PubMed: 37171131
- DOI: https://doi.org/10.1002/prot.26498
- Primary Citation of Related Structures:
8H56 - PubMed Abstract:
Porcine circovirus type 2 (PCV2) can cause porcine circovirus-associated disease (PCVAD), which causes significant economic losses to the global pig industry annually. There are no effective antiviral drugs used to control and treat PCV2, and prevention is mainly obtained through vaccination. PCV2 genome replicates through the rolling circle replication (RCR) mechanism involving Rep and Rep', so analyzing the holistic structure of Rep and Rep' will help us better understand the replication process of PCV2. However, there are no reports on the integral structure of Rep' and Rep, which seriously hinders the research of the viral replication. By using the x-ray diffraction method, the structure of the Rep' dimer was resolved by us in this study. Structural analysis revealed that Rep' is a dimer formed by the interaction of the C-terminal domain. The two Rep' form a positively charged groove, which may play an essential role in the viral binding of dsDNA. Together, this study help to understand the replication process of the virus and may also provide new insights into the development of antiviral drugs.
- State Key Laboratory of Agricultural Microbiology, Huazhong Agricultural University, Wuhan, China.
Organizational Affiliation: