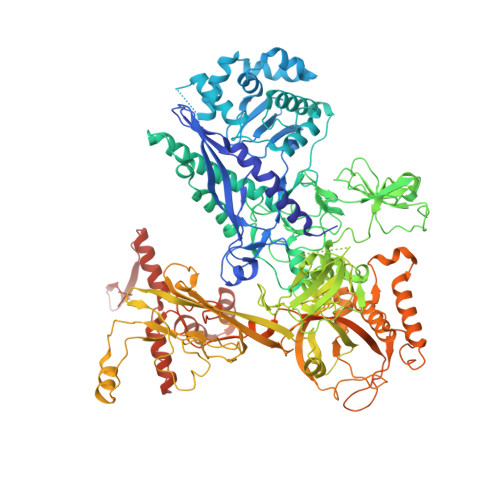
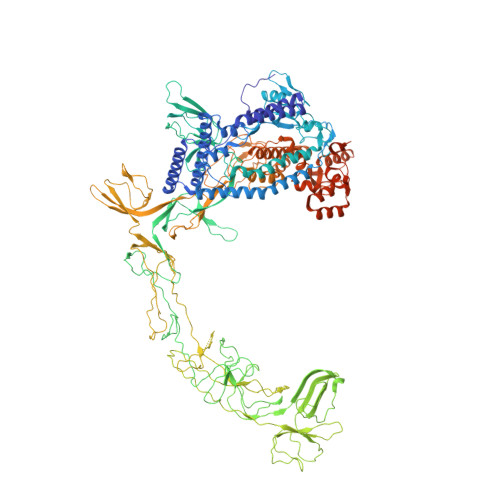
An SI3-sigma arch stabilizes cyanobacteria transcription initiation complex.
Shen, L., Lai, G., You, L., Shi, J., Wu, X., Puiu, M., Gu, Z., Feng, Y., Yuzenkova, Y., Zhang, Y.(2023) Proc Natl Acad Sci U S A 120: e2219290120-e2219290120
- PubMed: 37036976
- DOI: https://doi.org/10.1073/pnas.2219290120
- Primary Citation of Related Structures:
8GZG, 8GZH, 8H02 - PubMed Abstract:
Multisubunit RNA polymerases (RNAPs) associate with initiation factors (σ in bacteria) to start transcription. The σ factors are responsible for recognizing and unwinding promoter DNA in all bacterial RNAPs. Here, we report two cryo-EM structures of cyanobacterial transcription initiation complexes at near-atomic resolutions. The structures show that cyanobacterial RNAP forms an "SI3-σ" arch interaction between domain 2 of σ A (σ 2 ) and sequence insertion 3 (SI3) in the mobile catalytic domain Trigger Loop (TL). The "SI3-σ" arch facilitates transcription initiation from promoters of different classes through sealing the main cleft and thereby stabilizing the RNAP-promoter DNA open complex. Disruption of the "SI3-σ" arch disturbs cyanobacteria growth and stress response. Our study reports the structure of cyanobacterial RNAP and a unique mechanism for its transcription initiation. Our data suggest functional plasticity of SI3 and provide the foundation for further research into cyanobacterial and chloroplast transcription.
- Key Laboratory of Synthetic Biology, Chinese Academy of Sciences Center for Excellence in Molecular Plant Sciences, Shanghai Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai 200032, China.
Organizational Affiliation: