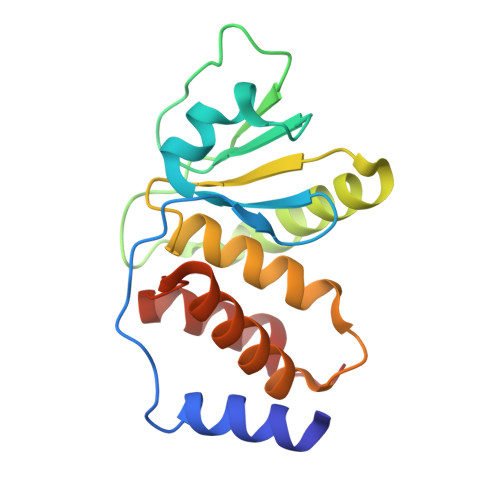
Crystal structure of monkeypox H1 phosphatase, an antiviral drug target.
Cui, W., Huang, H., Duan, Y., Luo, Z., Wang, H., Zhang, T., Nguyen, H.C., Shen, W., Su, D., Li, X., Ji, X., Yang, H., Wang, W.(2023) Protein Cell 14: 469-472
- PubMed: 37285262
- DOI: https://doi.org/10.1093/procel/pwac051
- Primary Citation of Related Structures:
8GZ4 - Institute of Life Sciences, Chongqing Medical University, Chongqing 400016, China.
Organizational Affiliation: