High-resolution structure of the cemiplimab Fab in complex with PD-1
Heo, Y.-S., Jeong, T.J.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| heavy chain | A, D [auth H] | 229 | Homo sapiens | Mutation(s): 0 | 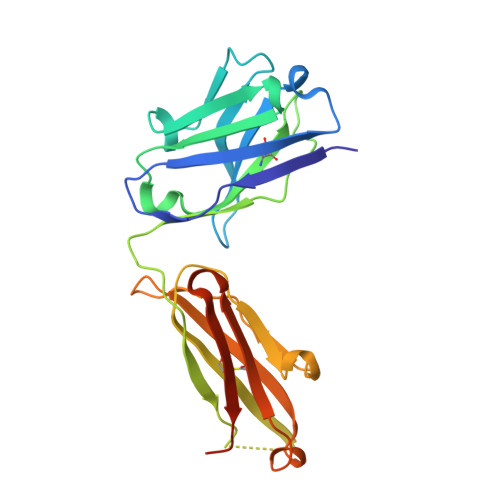 |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| light chain | B, E [auth L] | 214 | Homo sapiens | Mutation(s): 0 | 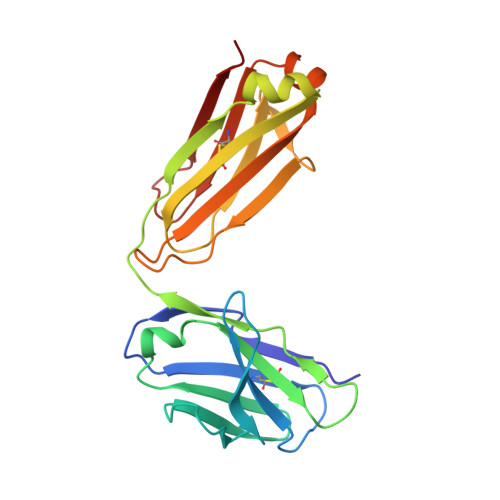 |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Programmed cell death protein 1 | C [auth Q], F [auth P] | 125 | Homo sapiens | Mutation(s): 0 Gene Names: PDCD1, PD1 | 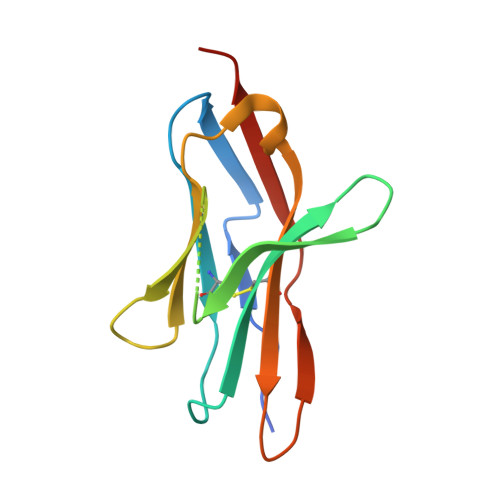 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q15116 (Homo sapiens) Explore Q15116 Go to UniProtKB: Q15116 | |||||
PHAROS: Q15116 GTEx: ENSG00000188389 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q15116 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 55.06 | α = 105.73 |
| b = 62.96 | β = 98 |
| c = 81.29 | γ = 92.44 |
| Software Name | Purpose |
|---|---|
| XDS | data scaling |
| PHENIX | refinement |
| PDB_EXTRACT | data extraction |
| XDS | data reduction |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Research Foundation (NRF, Korea) | Korea, Republic Of | -- |