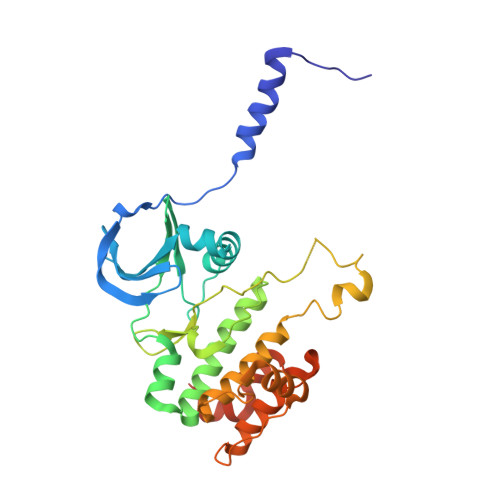
Structural basis for CEP192-mediated regulation of centrosomal AURKA.
Park, J.G., Jeon, H., Shin, S., Song, C., Lee, H., Kim, N.K., Kim, E.E., Hwang, K.Y., Lee, B.J., Lee, I.G.(2023) Sci Adv 9: eadf8582-eadf8582
- PubMed: 37083534
- DOI: https://doi.org/10.1126/sciadv.adf8582
- Primary Citation of Related Structures:
8GUW - PubMed Abstract:
Aurora kinase A (AURKA) performs critical functions in mitosis. Thus, the activity and subcellular localization of AURKA are tightly regulated and depend on diverse factors including interactions with the multiple binding cofactors. How these different cofactors regulate AURKA to elicit different levels of activity at distinct subcellular locations and times is poorly understood. Here, we identified a conserved region of CEP192, the major cofactor of AURKA, that mediates the interaction with AURKA. Quantitative binding studies were performed to map the interactions of a conserved helix (Helix-1) within CEP192. The crystal structure of Helix-1 bound to AURKA revealed a distinct binding site that is different from other cofactor proteins such as TPX2. Inhibiting the interaction between Helix-1 and AURKA in cells led to the mitotic defects, demonstrating the importance of the interaction. Collectively, we revealed a structural basis for the CEP192-mediated AURKA regulation at the centrosome, which is distinct from TPX2-mediated regulation on the spindle microtubule.
- Biomedical Research Division, Korea Institute of Science and Technology, Seoul 02792, South Korea.
Organizational Affiliation: