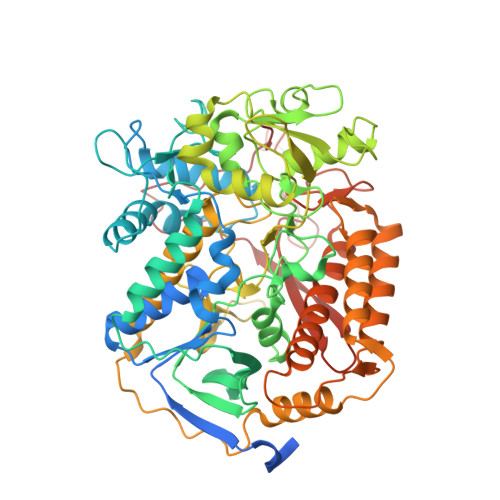
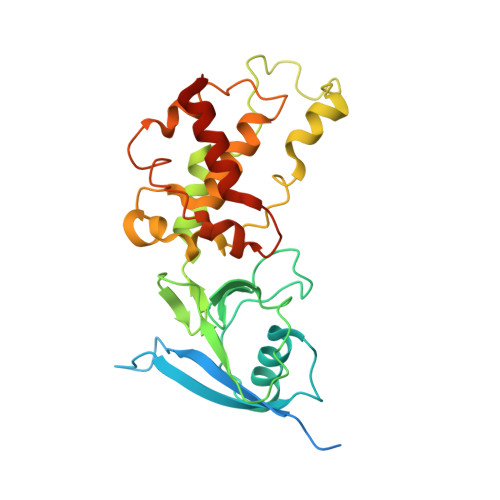
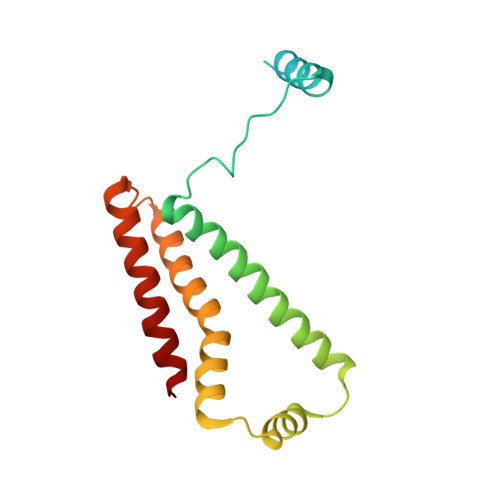
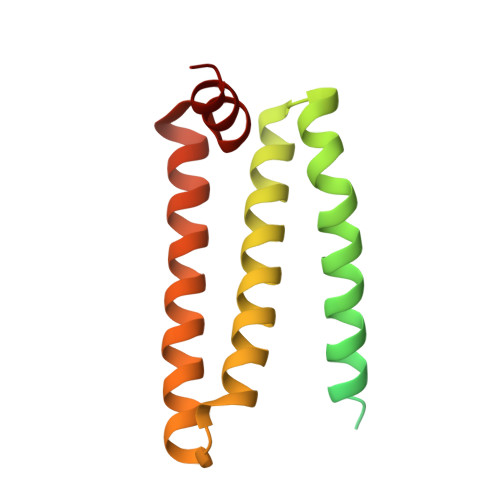
Structure of the human respiratory complex II.
Du, Z., Zhou, X., Lai, Y., Xu, J., Zhang, Y., Zhou, S., Feng, Z., Yu, L., Tang, Y., Wang, W., Yu, L., Tian, C., Ran, T., Chen, H., Guddat, L.W., Liu, F., Gao, Y., Rao, Z., Gong, H.(2023) Proc Natl Acad Sci U S A 120: e2216713120-e2216713120
- PubMed: 37098072
- DOI: https://doi.org/10.1073/pnas.2216713120
- Primary Citation of Related Structures:
8GS8 - PubMed Abstract:
Human complex II is a key protein complex that links two essential energy-producing processes: the tricarboxylic acid cycle and oxidative phosphorylation. Deficiencies due to mutagenesis have been shown to cause mitochondrial disease and some types of cancers. However, the structure of this complex is yet to be resolved, hindering a comprehensive understanding of the functional aspects of this molecular machine. Here, we have determined the structure of human complex II in the presence of ubiquinone at 2.86 Å resolution by cryoelectron microscopy, showing it comprises two water-soluble subunits, SDHA and SDHB, and two membrane-spanning subunits, SDHC and SDHD. This structure allows us to propose a route for electron transfer. In addition, clinically relevant mutations are mapped onto the structure. This mapping provides a molecular understanding to explain why these variants have the potential to produce disease.
- State Key Laboratory of Medicinal Chemical Biology, Frontiers Science Center for Cell Responses, College of Life Sciences, Nankai University, Tianjin 300353, China.
Organizational Affiliation: