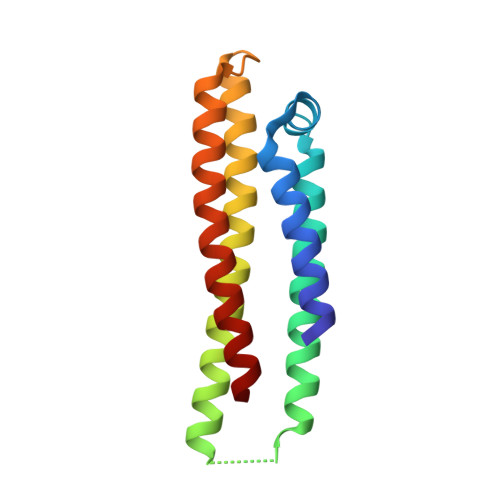
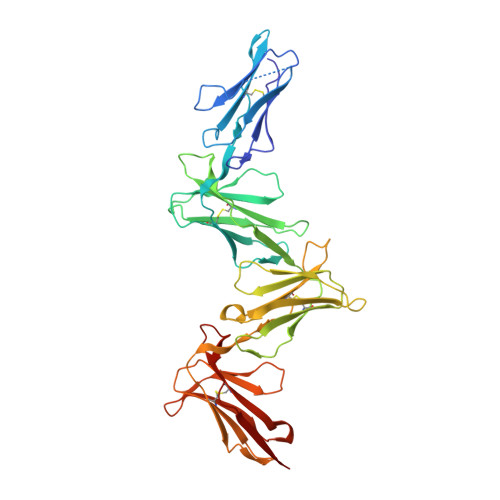
LilrB3 is a putative cell surface receptor of APOE4.
Zhou, J., Wang, Y., Huang, G., Yang, M., Zhu, Y., Jin, C., Jing, D., Ji, K., Shi, Y.(2023) Cell Res 33: 116-130
- PubMed: 36588123
- DOI: https://doi.org/10.1038/s41422-022-00759-y
- Primary Citation of Related Structures:
8GRX - PubMed Abstract:
The three isoforms of apolipoprotein E (APOE2, APOE3, and APOE4) only differ in two amino acid positions but exert quite different immunomodulatory effects. The underlying mechanism of such APOE isoform dependence remains enigmatic. Here we demonstrate that APOE4, but not APOE2, specifically interacts with the leukocyte immunoglobulin-like receptor B3 (LilrB3). Two discrete immunoglobin-like domains of the LilrB3 extracellular domain (ECD) recognize a positively charged surface patch on the N-terminal domain (NTD) of APOE4. The atomic structure reveals how two APOE4 molecules specifically engage two LilrB3 molecules, bringing their intracellular signaling motifs into close proximity through formation of a hetero-tetrameric complex. Consistent with our biochemical and structural analyses, APOE4, but not APOE2, activates human microglia cells (HMC3) into a pro-inflammatory state in a LilrB3-dependent manner. Together, our study identifies LilrB3 as a putative immune cell surface receptor for APOE4, but not APOE2, and may have implications for understanding the biological functions as well as disease relevance of the APOE isoforms.
- Key Laboratory of Structural Biology of Zhejiang Province, School of Life Sciences, Westlake University; Institute of Biology, Westlake Institute for Advanced Study, Hangzhou, Zhejiang, China. zhoujiayao@westlake.edu.cn.
Organizational Affiliation: