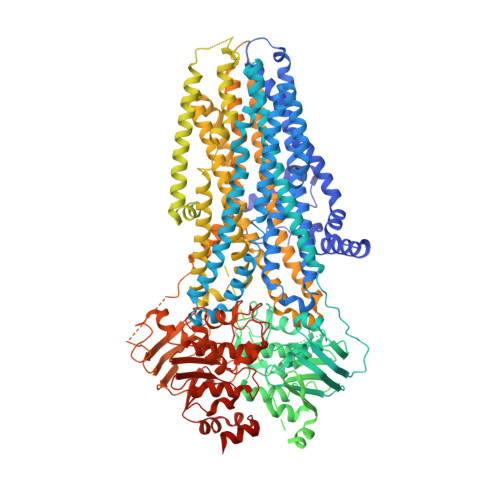
Structure-based discovery of CFTR potentiators and inhibitors.
Liu, F., Kaplan, A.L., Levring, J., Einsiedel, J., Tiedt, S., Distler, K., Omattage, N.S., Kondratov, I.S., Moroz, Y.S., Pietz, H.L., Irwin, J.J., Gmeiner, P., Shoichet, B.K., Chen, J.(2024) Cell 187: 3712-3725.e34
- PubMed: 38810646
- DOI: https://doi.org/10.1016/j.cell.2024.04.046
- Primary Citation of Related Structures:
8GLS - PubMed Abstract:
The cystic fibrosis transmembrane conductance regulator (CFTR) is a crucial ion channel whose loss of function leads to cystic fibrosis, whereas its hyperactivation leads to secretory diarrhea. Small molecules that improve CFTR folding (correctors) or function (potentiators) are clinically available. However, the only potentiator, ivacaftor, has suboptimal pharmacokinetics and inhibitors have yet to be clinically developed. Here, we combine molecular docking, electrophysiology, cryo-EM, and medicinal chemistry to identify CFTR modulators. We docked ∼155 million molecules into the potentiator site on CFTR, synthesized 53 test ligands, and used structure-based optimization to identify candidate modulators. This approach uncovered mid-nanomolar potentiators, as well as inhibitors, that bind to the same allosteric site. These molecules represent potential leads for the development of more effective drugs for cystic fibrosis and secretory diarrhea, demonstrating the feasibility of large-scale docking for ion channel drug discovery.
- Laboratory of Membrane Biology and Biophysics, The Rockefeller University, New York, NY 10065, USA; Department of Pharmaceutical Chemistry, University of California, San Francisco, San Francisco, CA 94143, USA.
Organizational Affiliation: