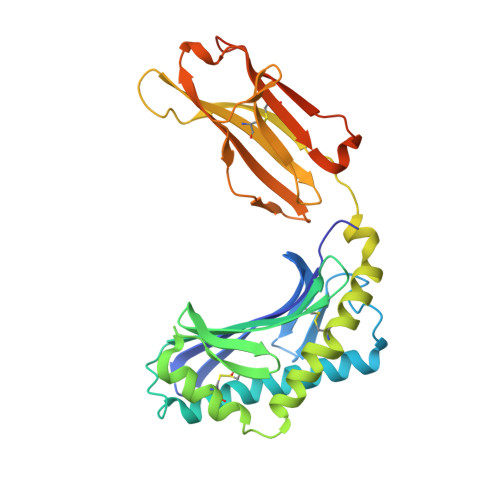
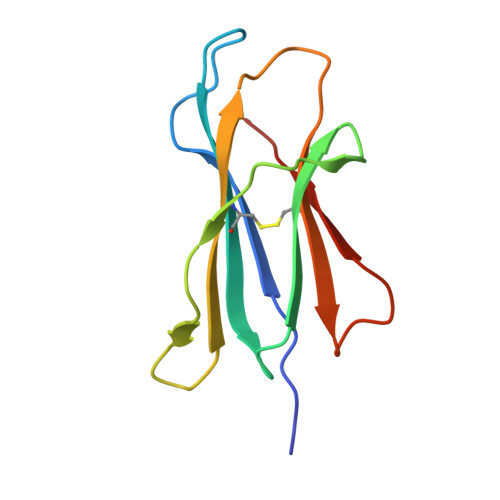
CD1 lipidomes reveal lipid-binding motifs and size-based antigen-display mechanisms.
Huang, S., Shahine, A., Cheng, T.Y., Chen, Y.L., Ng, S.W., Balaji, G.R., Farquhar, R., Gras, S., Hardman, C.S., Altman, J.D., Tahiri, N., Minnaard, A.J., Ogg, G.S., Mayfield, J.A., Rossjohn, J., Moody, D.B.(2023) Cell 186: 4583-4596.e13
- PubMed: 37725977
- DOI: https://doi.org/10.1016/j.cell.2023.08.022
- Primary Citation of Related Structures:
8GLE, 8GLF, 8GLG, 8GLH, 8GLI - PubMed Abstract:
The CD1 system binds lipid antigens for display to T cells. Here, we solved lipidomes for the four human CD1 antigen-presenting molecules, providing a map of self-lipid display. Answering a basic question, the detection of >2,000 CD1-lipid complexes demonstrates broad presentation of self-sphingolipids and phospholipids. Whereas peptide antigens are chemically processed, many lipids are presented in an unaltered form. However, each type of CD1 protein differentially edits the self-lipidome to show distinct capture motifs based on lipid length and chemical composition, suggesting general antigen display mechanisms. For CD1a and CD1d, lipid size matches the CD1 cleft volume. CD1c cleft size is more variable, and CD1b is the outlier, where ligands and clefts show an extreme size mismatch that is explained by uniformly seating two small lipids in one cleft. Furthermore, the list of compounds that comprise the integrated CD1 lipidome supports the ongoing discovery of lipid blockers and antigens for T cells.
- Division of Rheumatology, Immunity and Inflammation, Brigham and Women's Hospital, Harvard Medical School, Boston, MA 02115, USA. Electronic address: shouxiong.huang@uc.edu.
Organizational Affiliation: