Characterization of a Potent and Orally Bioavailable Lys-Covalent Inhibitor of Apoptosis Protein (IAP) Antagonist.
Udompholkul, P., Garza-Granados, A., Alboreggia, G., Baggio, C., McGuire, J., Pegan, S.D., Pellecchia, M.(2023) J Med Chem 66: 8159-8169
- PubMed: 37262387
- DOI: https://doi.org/10.1021/acs.jmedchem.3c00467
- Primary Citation of Related Structures:
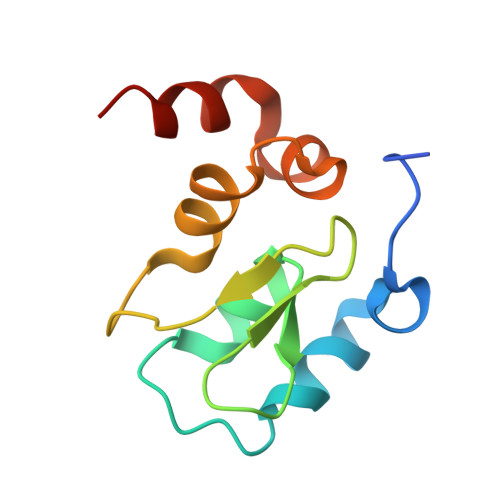
8GH7 - PubMed Abstract:
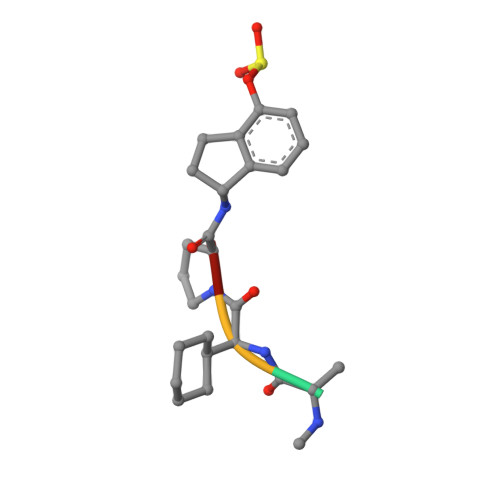
We have recently reported on the use of aryl-fluorosulfates in designing water- and plasma-stable agents that covalently target Lys, Tyr, or His residues in the BIR3 domain of the inhibitor of the apoptosis protein (IAP) family. Here, we report further structural, cellular, and pharmacological characterizations of this agent, including the high-resolution structure of the complex between the Lys-covalent agent and its target, the BIR3 domain of X-linked IAP (XIAP). We also compared the cellular efficacy of the agent in two-dimensional (2D) and three-dimensional (3D) cell cultures, side by side with the clinical candidate reversible IAP inhibitor LCL161. Finally, in vivo pharmacokinetic studies indicated that the agent was long-lived and orally bioavailable. Collectively our data further corroborate that aryl-fluorosulfates, when incorporated correctly in a ligand, can result in Lys-covalent agents with pharmacodynamic and pharmacokinetic properties that warrant their use in the design of pharmacological probes or even therapeutics.
- Division of Biomedical Sciences, School of Medicine, University of California, Riverside, 900 University Avenue, Riverside, California 92521, United States.
Organizational Affiliation: