Structural insights into functional properties of the oxidized form of cytochrome c oxidase.
Ishigami, I., Sierra, R.G., Su, Z., Peck, A., Wang, C., Poitevin, F., Lisova, S., Hayes, B., Moss 3rd, F.R., Boutet, S., Sublett, R.E., Yoon, C.H., Yeh, S.R., Rousseau, D.L.(2023) Nat Commun 14: 5752-5752
- PubMed: 37717031
- DOI: https://doi.org/10.1038/s41467-023-41533-x
- Primary Citation of Related Structures:
8GCQ - PubMed Abstract:
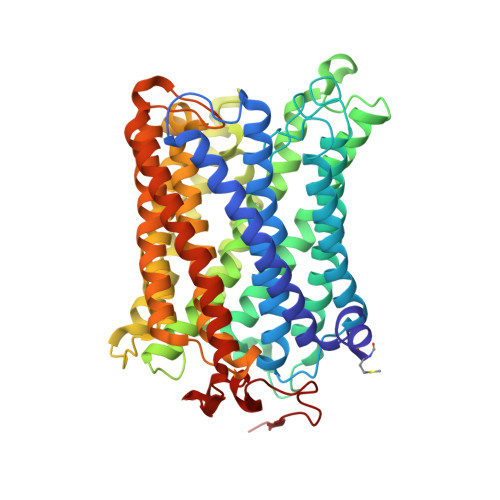
Cytochrome c oxidase (CcO) is an essential enzyme in mitochondrial and bacterial respiration. It catalyzes the four-electron reduction of molecular oxygen to water and harnesses the chemical energy to translocate four protons across biological membranes. The turnover of the CcO reaction involves an oxidative phase, in which the reduced enzyme (R) is oxidized to the metastable O H state, and a reductive phase, in which O H is reduced back to the R state. During each phase, two protons are translocated across the membrane. However, if O H is allowed to relax to the resting oxidized state (O), a redox equivalent to O H , its subsequent reduction to R is incapable of driving proton translocation. Here, with resonance Raman spectroscopy and serial femtosecond X-ray crystallography (SFX), we show that the heme a 3 iron and Cu B in the active site of the O state, like those in the O H state, are coordinated by a hydroxide ion and a water molecule, respectively. However, Y244, critical for the oxygen reduction chemistry, is in the neutral protonated form, which distinguishes O from O H , where Y244 is in the deprotonated tyrosinate form. These structural characteristics of O provide insights into the proton translocation mechanism of CcO.
- Department of Biochemistry, Albert Einstein College of Medicine, Bronx, NY, 10461, USA.
Organizational Affiliation: