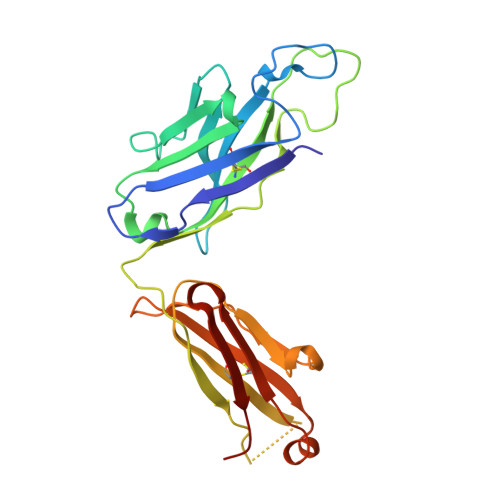
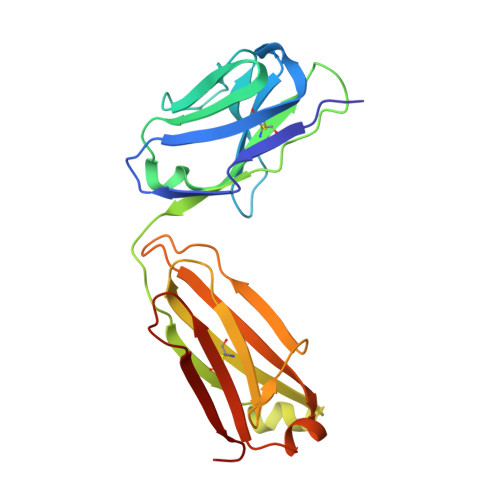
Antigen pressure from two founder viruses induces multiple insertions at a single antibody position to generate broadly neutralizing HIV antibodies.
Joyce, C., Murrell, S., Murrell, B., Omorodion, O., Ver, L.S., Carrico, N., Bastidas, R., Nedellec, R., Bick, M., Woehl, J., Zhao, F., Burns, A., Barman, S., Appel, M., Ramos, A., Wickramasinghe, L., Eren, K., Vollbrecht, T., Smith, D.M., Kosakovsky Pond, S.L., McBride, R., Worth, C., Batista, F., Sok, D., Poignard, P., Briney, B., Wilson, I.A., Landais, E., Burton, D.R.(2023) PLoS Pathog 19: e1011416-e1011416
- PubMed: 37384622
- DOI: https://doi.org/10.1371/journal.ppat.1011416
- Primary Citation of Related Structures:
8GBV, 8GBW, 8GBX, 8GBY, 8GBZ, 8GC0, 8GC1 - PubMed Abstract:
Vaccination strategies aimed at maturing broadly neutralizing antibodies (bnAbs) from naïve precursors are hindered by unusual features that characterize these Abs, including insertions and deletions (indels). Longitudinal studies of natural HIV infection cases shed light on the complex processes underlying bnAb development and have suggested a role for superinfection as a potential enhancer of neutralization breadth. Here we describe the development of a potent bnAb lineage that was elicited by two founder viruses to inform vaccine design. The V3-glycan targeting bnAb lineage (PC39-1) was isolated from subtype C-infected IAVI Protocol C elite neutralizer, donor PC39, and is defined by the presence of multiple independent insertions in CDRH1 that range from 1-11 amino acids in length. Memory B cell members of this lineage are predominantly atypical in phenotype yet also span the class-switched and antibody-secreting cell compartments. Development of neutralization breadth occurred concomitantly with extensive recombination between founder viruses before each virus separated into two distinct population "arms" that evolved independently to escape the PC39-1 lineage. Ab crystal structures show an extended CDRH1 that can help stabilize the CDRH3. Overall, these findings suggest that early exposure of the humoral system to multiple related Env molecules could promote the induction of bnAbs by focusing Ab responses to conserved epitopes.
- Department of Immunology and Microbiology, The Scripps Research Institute, La Jolla, California, United States of America.
Organizational Affiliation: