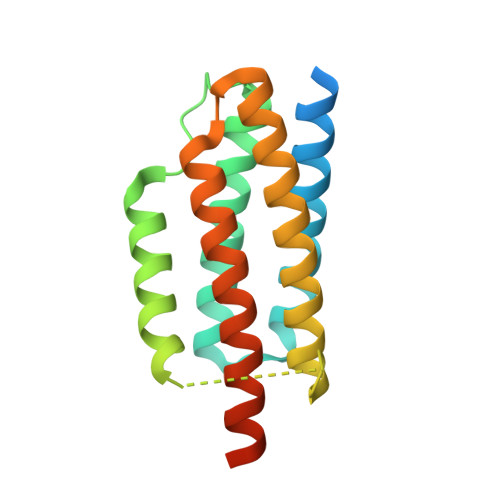
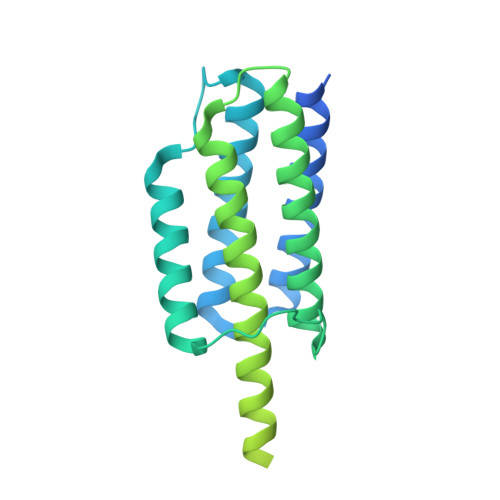
Cryo-EM structure determination of small therapeutic protein targets at 3 angstrom -resolution using a rigid imaging scaffold.
Castells-Graells, R., Meador, K., Arbing, M.A., Sawaya, M.R., Gee, M., Cascio, D., Gleave, E., Debreczeni, J.E., Breed, J., Leopold, K., Patel, A., Jahagirdar, D., Lyons, B., Subramaniam, S., Phillips, C., Yeates, T.O.(2023) Proc Natl Acad Sci U S A 120: e2305494120-e2305494120
- PubMed: 37669364
- DOI: https://doi.org/10.1073/pnas.2305494120
- Primary Citation of Related Structures:
8G3K, 8G42, 8G47, 8G4E, 8G4F, 8G4H - PubMed Abstract:
Cryoelectron microscopy (Cryo-EM) has enabled structural determination of proteins larger than about 50 kDa, including many intractable by any other method, but it has largely failed for smaller proteins. Here, we obtain structures of small proteins by binding them to a rigid molecular scaffold based on a designed protein cage, revealing atomic details at resolutions reaching 2.9 Å. We apply this system to the key cancer signaling protein KRAS (19 kDa in size), obtaining four structures of oncogenic mutational variants by cryo-EM. Importantly, a structure for the key G12C mutant bound to an inhibitor drug (AMG510) reveals significant conformational differences compared to prior data in the crystalline state. The findings highlight the promise of cryo-EM scaffolds for advancing the design of drug molecules against small therapeutic protein targets in cancer and other human diseases.
- Department of Energy, Institute for Genomics and Proteomics, University of California, Los Angeles, CA 90095.
Organizational Affiliation: