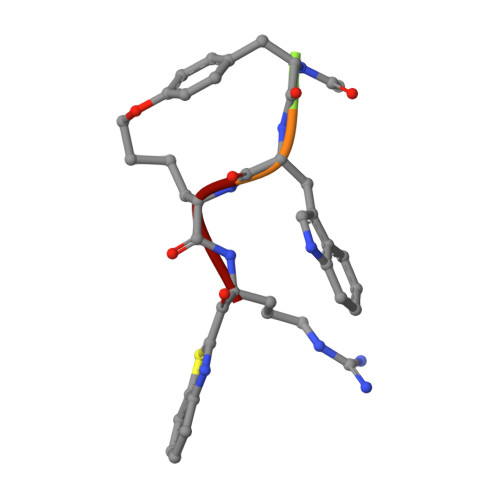
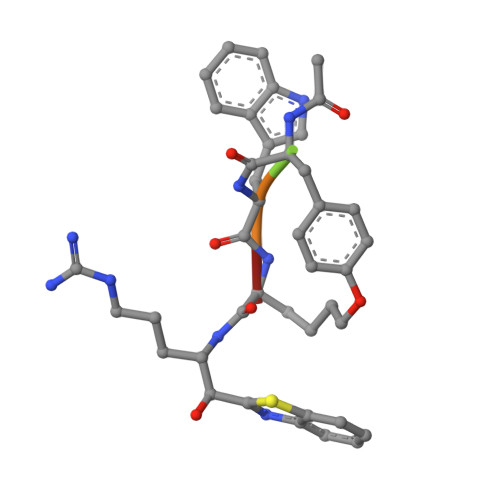
Mechanism-Based Macrocyclic Inhibitors of Serine Proteases.
Damalanka, V.C., Banas, V., De Bona, P., Kashipathy, M.M., Battaile, K., Lovell, S., Janetka, J.W.(2024) J Med Chem 67: 4833-4854
- PubMed: 38477709
- DOI: https://doi.org/10.1021/acs.jmedchem.3c02388
- Primary Citation of Related Structures:
8G1W - PubMed Abstract:
Protease inhibitor drug discovery is challenged by the lack of cellular and oral permeability, selectivity, metabolic stability, and rapid clearance of peptides. Here, we describe the rational design, synthesis, and evaluation of peptidomimetic side-chain-cyclized macrocycles which we converted into covalent serine protease inhibitors with the addition of an electrophilic ketone warhead. We have identified potent and selective inhibitors of TMPRSS2, matriptase, hepsin, and HGFA and demonstrated their improved protease selectivity, metabolic stability, and pharmacokinetic (PK) properties. We obtained an X-ray crystal structure of phenyl ether-cyclized tripeptide VD4162 ( 8b ) bound to matriptase, revealing an unexpected binding conformation. Cyclic biphenyl ether VD5123 ( 11 ) displayed the best PK properties in mice with a half-life of 4.5 h and compound exposure beyond 24 h. These new cyclic tripeptide scaffolds can be used as easily modifiable templates providing a new strategy to overcoming the obstacles presented by linear acyclic peptides in protease inhibitor drug discovery.
- Department of Biochemistry & Molecular Biophysics, Washington University School of Medicine, Saint Louis, Missouri 63110, United States.
Organizational Affiliation: