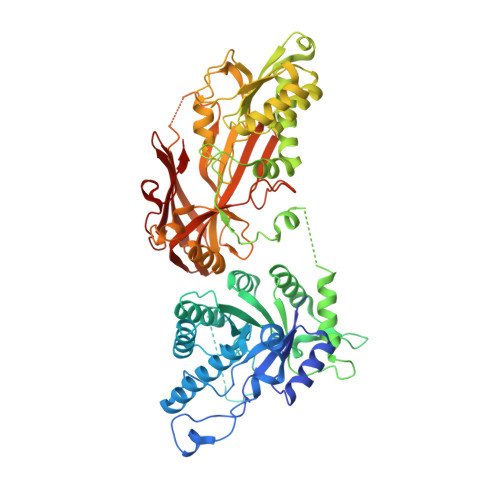
In vitro methylation of the U7 snRNP subunits Lsm11 and SmE by the PRMT5/MEP50/pICln methylosome.
Yang, X.C., Desotell, A., Lin, M.H., Paige, A.S., Malinowska, A., Sun, Y., Aik, W.S., Dadlez, M., Tong, L., Dominski, Z.(2023) RNA 29: 1673-1690
- PubMed: 37562960
- DOI: https://doi.org/10.1261/rna.079709.123
- Primary Citation of Related Structures:
8G1U - PubMed Abstract:
U7 snRNP is a multisubunit endonuclease required for 3' end processing of metazoan replication-dependent histone pre-mRNAs. In contrast to the spliceosomal snRNPs, U7 snRNP lacks the Sm subunits D1 and D2 and instead contains two related proteins, Lsm10 and Lsm11. The remaining five subunits of the U7 heptameric Sm ring, SmE, F, G, B, and D3, are shared with the spliceosomal snRNPs. The pathway that assembles the unique ring of U7 snRNP is unknown. Here, we show that a heterodimer of Lsm10 and Lsm11 tightly interacts with the methylosome, a complex of the arginine methyltransferase PRMT5, MEP50, and pICln known to methylate arginines in the carboxy-terminal regions of the Sm proteins B, D1, and D3 during the spliceosomal Sm ring assembly. Both biochemical and cryo-EM structural studies demonstrate that the interaction is mediated by PRMT5, which binds and methylates two arginine residues in the amino-terminal region of Lsm11. Surprisingly, PRMT5 also methylates an amino-terminal arginine in SmE, a subunit that does not undergo this type of modification during the biogenesis of the spliceosomal snRNPs. An intriguing possibility is that the unique methylation pattern of Lsm11 and SmE plays a vital role in the assembly of the U7 snRNP.
- Integrative Program for Biological and Genome Sciences, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina 27599, USA.
Organizational Affiliation: