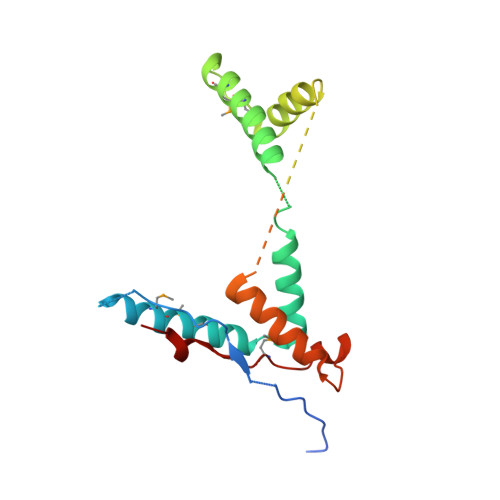
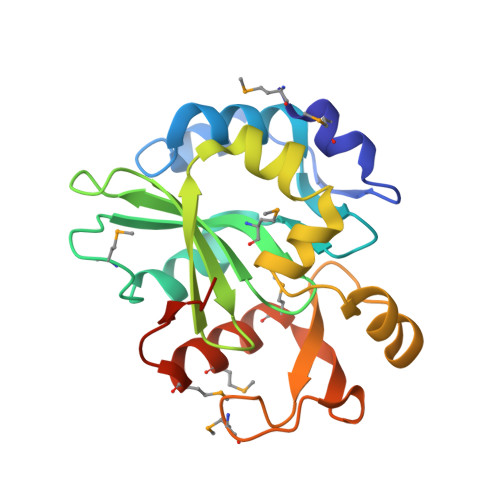
Structural disruption of Ntox15 nuclease effector domains by immunity proteins protects against type VI secretion system intoxication in Bacteroidales.
Bosch, D.E., Abbasian, R., Parajuli, B., Peterson, S.B., Mougous, J.D.(2023) mBio 14: e0103923-e0103923
- PubMed: 37345922
- DOI: https://doi.org/10.1128/mbio.01039-23
- Primary Citation of Related Structures:
8FZY, 8FZZ, 8G0K - PubMed Abstract:
Bacteroidales use type VI secretion systems (T6SS) to competitively colonize and persist in the colon. We identify a horizontally transferred T6SS with Ntox15 family nuclease effector (Tde1) that mediates interbacterial antagonism among Bacteroidales, including several derived from a single human donor. Expression of cognate (Tdi1) or orphan immunity proteins in acquired interbacterial defense systems protects against Tde1-dependent attack. We find that immunity protein interaction induces a large effector conformational change in Tde nucleases, disrupting the active site and altering the DNA-binding site. Crystallographic snapshots of isolated Tde1, the Tde1/Tdi1 complex, and homologs from Phocaeicola vulgatus (Tde2/Tdi2) illustrate a conserved mechanism of immunity inserting into the central core of Tde, splitting the nuclease fold into two subdomains. The Tde/Tdi interface and immunity mechanism are distinct from all other polymorphic toxin-immunity interactions of known structure. Bacteroidales abundance has been linked to inflammatory bowel disease activity in prior studies, and we demonstrate that Tde and T6SS structural genes are each enriched in fecal metagenomes from ulcerative colitis subjects. Genetically mobile Tde1-encoding T6SS in Bacteroidales mediate competitive growth and may be involved in inflammatory bowel disease. Broad immunity is conferred by Tdi1 homologs through a fold-disrupting mechanism unique among polymorphic effector-immunity pairs of known structure. IMPORTANCE Bacteroidales are related to inflammatory bowel disease severity and progression. We identify type VI secretion system (T6SS) nuclease effectors (Tde) which are enriched in ulcerative colitis and horizontally transferred on mobile genetic elements. Tde-encoding T6SSs mediate interbacterial competition. Orphan and cognate immunity proteins (Tdi) prevent intoxication by multiple Tde through a new mechanism among polymorphic toxin systems. Tdi inserts into the effector central core, splitting Ntox15 into two subdomains and disrupting the active site. This mechanism may allow for evolutionary diversification of the Tde/Tdi interface as observed in colicin nuclease-immunity interactions, promoting broad neutralization of Tde by orphan Tdi. Tde-dependent T6SS interbacterial antagonism may contribute to Bacteroidales diversity in the context of ulcerative colitis.
- Department of Pathology, Carver College of Medicine, University of Iowa , Iowa City, Iowa, USA.
Organizational Affiliation: