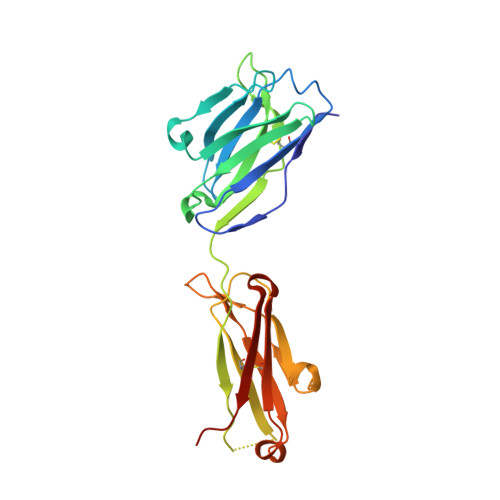
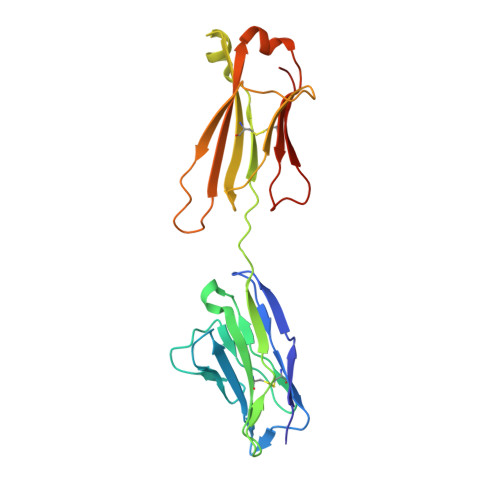
CryoEM structure of a therapeutic antibody (favezelimab) bound to human LAG3 determined using a bivalent Fab as fiducial marker.
Mishra, A.K., Shahid, S., Karade, S.S., Agnihotri, P., Kolesnikov, A., Hasan, S.S., Mariuzza, R.A.(2023) Structure 31: 1149
- PubMed: 37619561
- DOI: https://doi.org/10.1016/j.str.2023.07.013
- Primary Citation of Related Structures:
6WKM, 8FWH, 8SO3, 8SR0 - PubMed Abstract:
Lymphocyte activation gene 3 protein (LAG3) is an inhibitory receptor that is upregulated on exhausted T cells in tumors. LAG3 is a major target for cancer immunotherapy with many anti-LAG3 antibodies in clinical trials. However, there is no structural information on the epitopes recognized by these antibodies. We determined the single-particle cryoEM structure of a therapeutic antibody (favezelimab) bound to LAG3 to 3.5 Å resolution, revealing that favezelimab targets the LAG3-binding site for MHC class II, its canonical ligand. The small size of the complex between the conventional (monovalent) Fab of favezelimab and LAG3 (∼100 kDa) presented a challenge for cryoEM. Accordingly, we engineered a bivalent version of Fab favezelimab that doubled the size of the Fab-LAG3 complex and conferred a highly identifiable shape to the complex that facilitated particle selection and orientation for image processing. This study establishes bivalent Fabs as new fiducial markers for cryoEM analysis of small proteins.
- W.M. Keck Laboratory for Structural Biology, University of Maryland Institute for Bioscience and Biotechnology Research, Rockville, MD 20850, USA; Department of Cell Biology and Molecular Genetics, University of Maryland, College Park, MD 20742, USA.
Organizational Affiliation: