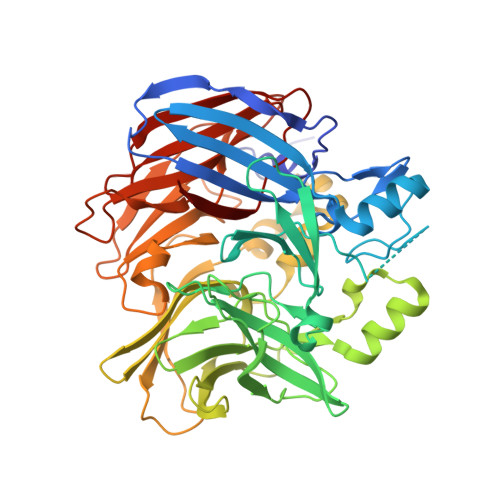
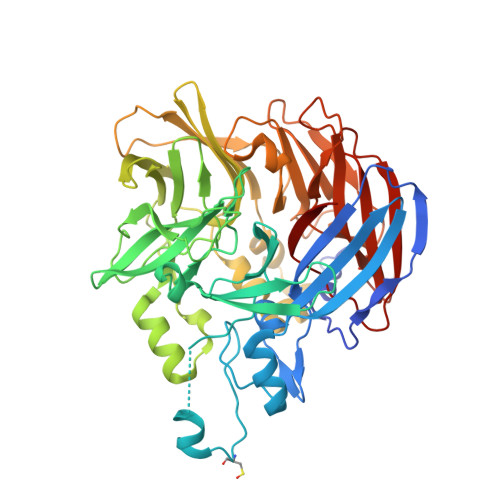
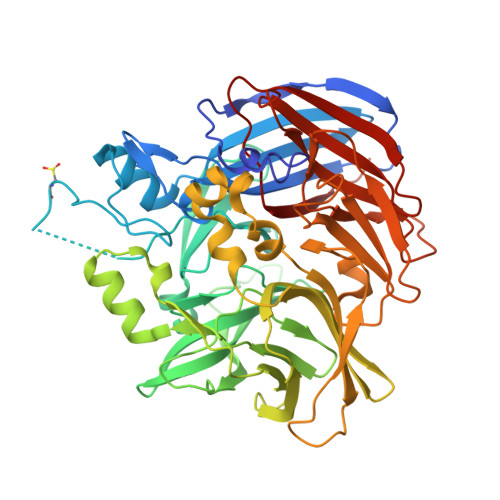
Carotenoid cleavage enzymes evolved convergently to generate the visual chromophore.
Solano, Y.J., Everett, M.P., Dang, K.S., Abueg, J., Kiser, P.D.(2024) Nat Chem Biol 20: 779-788
- PubMed: 38355721
- DOI: https://doi.org/10.1038/s41589-024-01554-z
- Primary Citation of Related Structures:
8FTY - PubMed Abstract:
The retinal light response in animals originates from the photoisomerization of an opsin-coupled 11-cis-retinaldehyde chromophore. This visual chromophore is enzymatically produced through the action of carotenoid cleavage dioxygenases. Vertebrates require two carotenoid cleavage dioxygenases, β-carotene oxygenase 1 and retinal pigment epithelium 65 (RPE65), to form 11-cis-retinaldehyde from carotenoid substrates, whereas invertebrates such as insects use a single enzyme known as Neither Inactivation Nor Afterpotential B (NinaB). RPE65 and NinaB couple trans-cis isomerization with hydrolysis and oxygenation, respectively, but the mechanistic relationship of their isomerase activities remains unknown. Here we report the structure of NinaB, revealing details of its active site architecture and mode of membrane binding. Structure-guided mutagenesis studies identify a residue cluster deep within the NinaB substrate-binding cleft that controls its isomerization activity. Our data demonstrate that isomerization activity is mediated by distinct active site regions in NinaB and RPE65-an evolutionary convergence that deepens our understanding of visual system diversity.
- Department of Physiology and Biophysics, University of California Irvine School of Medicine, Irvine, CA, USA.
Organizational Affiliation: