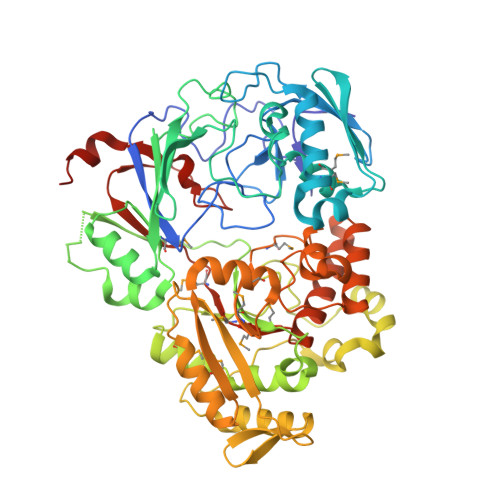
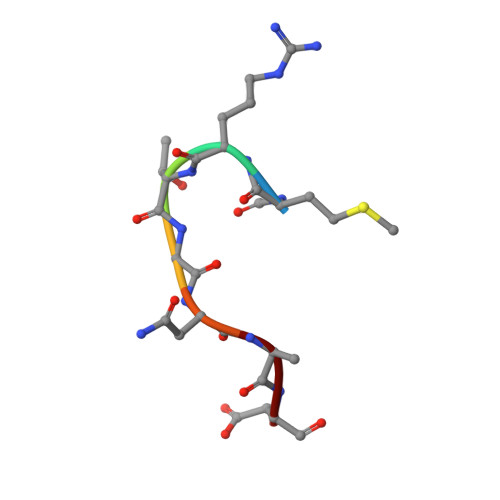
Trojan horse peptide conjugates remodel the activity spectrum of clinical antibiotics.
Luo, S., Li, X.R., Gong, X.T., Kulikovsky, A., Qu, F., Beis, K., Severinov, K., Dubiley, S., Feng, X., Dong, S.H., Nair, S.K.(2025) Proc Natl Acad Sci U S A 122: e2319483121-e2319483121
- PubMed: 39739799
- DOI: https://doi.org/10.1073/pnas.2319483121
- Primary Citation of Related Structures:
7Z6F, 8FSQ, 8FSR, 8FSS - PubMed Abstract:
Infections caused by gram-negative pathogens continue to be a major risk to human health because of the innate antibiotic resistance endowed by their unique cell membrane architecture. Nature has developed an elegant solution to target gram-negative strains, namely by conjugating toxic antibiotic warheads to a suitable carrier to facilitate the active import of the drug to a specific target organism. Microcin C7 (McC) is a Trojan horse peptide-conjugated antibiotic that specifically targets enterobacteria by exploiting active import through oligopeptide transport systems. Here, we characterize the molecular mechanism of McC recognition by YejA, the solute binding protein of the Escherichia coli oligopeptide transporter. Structure-guided mutational and functional analysis elucidates the determinants of substrate recognition. We demonstrate that the peptide carrier can serve as a passport for the entry of molecules that are otherwise not taken into E. coli cells. We show that peptide conjugation can remodel the antibiotic spectrum of clinically relevant parent compounds. Bioinformatics analysis reveals a broad distribution of YejA-like transporters in only the Proteobacteria, underscoring the potential for the development of Trojan horse antibiotics that are actively imported into such gram-negative bacteria.
- State Key Laboratory of Applied Organic Chemistry, College of Chemistry and Chemical Engineering, Lanzhou University, Lanzhou 730000, People's Republic of China.
Organizational Affiliation: