Comparative Study of Adenosine Analogs as Inhibitors of Protein Arginine Methyltransferases and a Clostridioides difficile- Specific DNA Adenine Methyltransferase.
Zhou, J., Deng, Y., Iyamu, I.D., Horton, J.R., Yu, D., Hajian, T., Vedadi, M., Rotili, D., Mai, A., Blumenthal, R.M., Zhang, X., Huang, R., Cheng, X.(2023) ACS Chem Biol 18: 734-745
- PubMed: 37082867
- DOI: https://doi.org/10.1021/acschembio.3c00035
- Primary Citation of Related Structures:
8FS1, 8FS2 - PubMed Abstract:
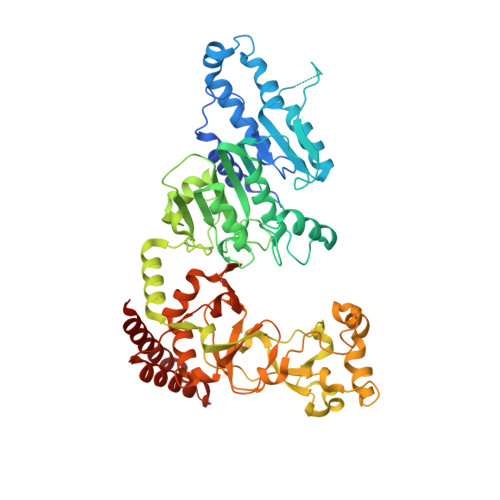
S -Adenosyl-l-methionine (SAM) analogs are adaptable tools for studying and therapeutically inhibiting SAM-dependent methyltransferases (MTases). Some MTases play significant roles in host-pathogen interactions, one of which is Clostridioides difficile -specific DNA adenine MTase (CamA). CamA is needed for efficient sporulation and alters persistence in the colon. To discover potent and selective CamA inhibitors, we explored modifications of the solvent-exposed edge of the SAM adenosine moiety. Starting from the two parental compounds ( 6e and 7 ), we designed an adenosine analog ( 11a ) carrying a 3-phenylpropyl moiety at the adenine N6-amino group, and a 3-(cyclohexylmethyl guanidine)-ethyl moiety at the sulfur atom off the ribose ring. Compound 11a (IC 50 = 0.15 μM) is 10× and 5× more potent against CamA than 6e and 7 , respectively. The structure of the CamA-DNA-inhibitor complex revealed that 11a adopts a U-shaped conformation, with the two branches folded toward each other, and the aliphatic and aromatic rings at the two ends interacting with one another. 11a occupies the entire hydrophobic surface (apparently unique to CamA) next to the adenosine binding site. Our work presents a hybrid knowledge-based and fragment-based approach to generating CamA inhibitors that would be chemical agents to examine the mechanism(s) of action and therapeutic potentials of CamA in C. difficile infection.
- Department of Epigenetics and Molecular Carcinogenesis, University of Texas MD Anderson Cancer Center, Houston, Texas 77030, United States.
Organizational Affiliation: