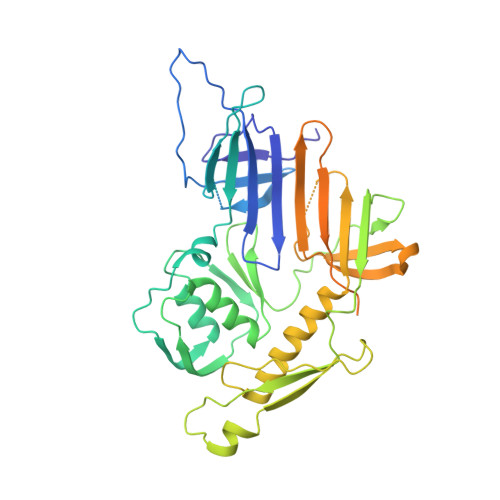
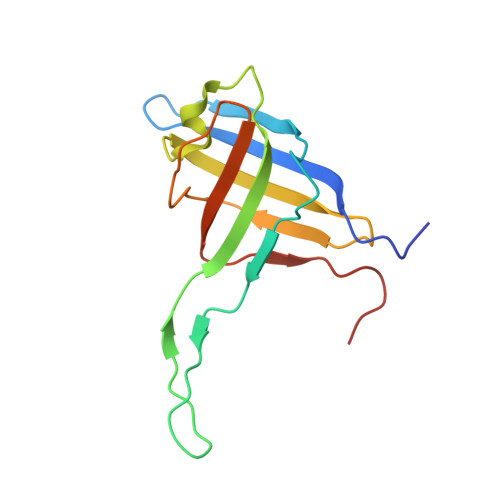
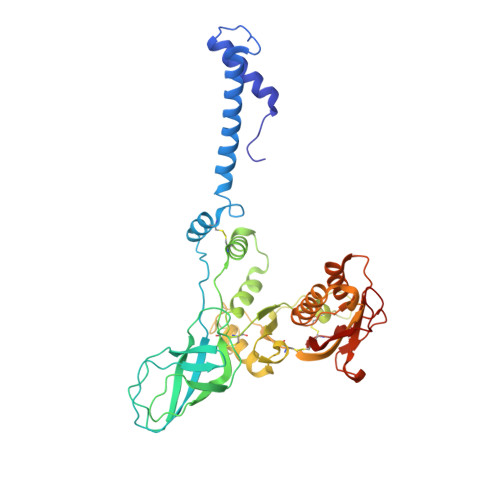
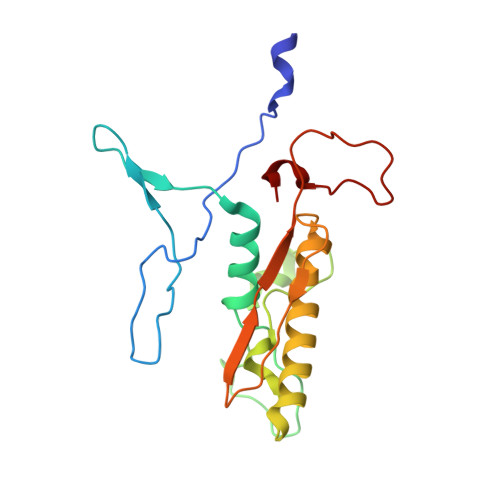
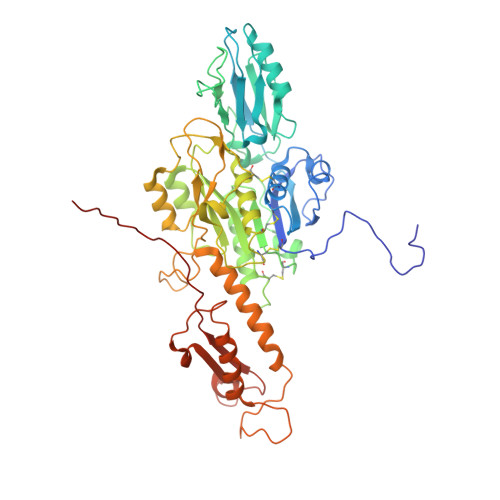
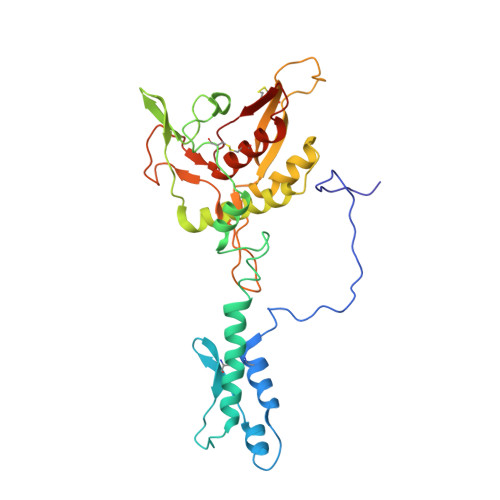
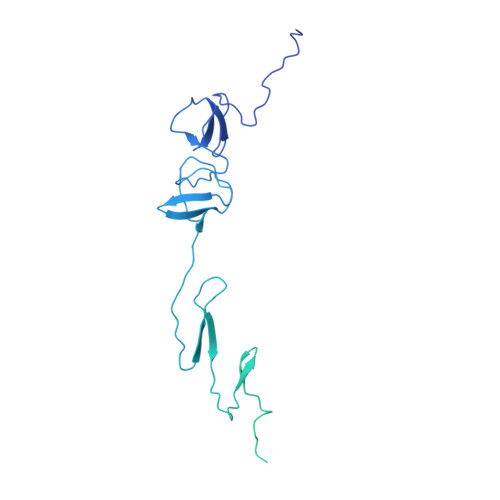
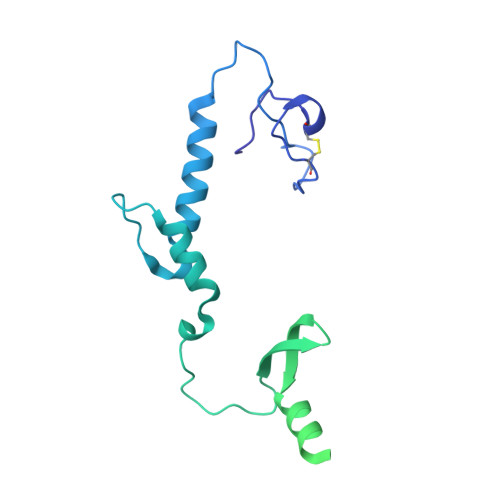
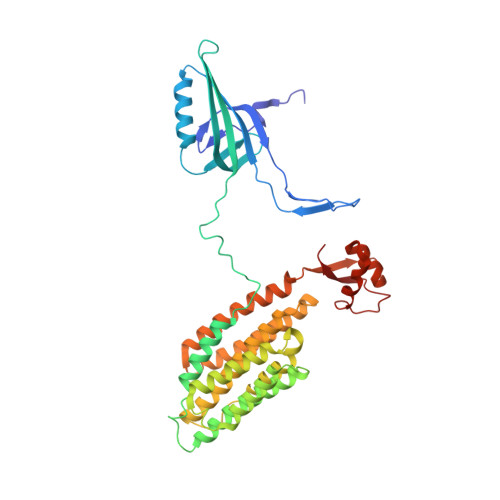
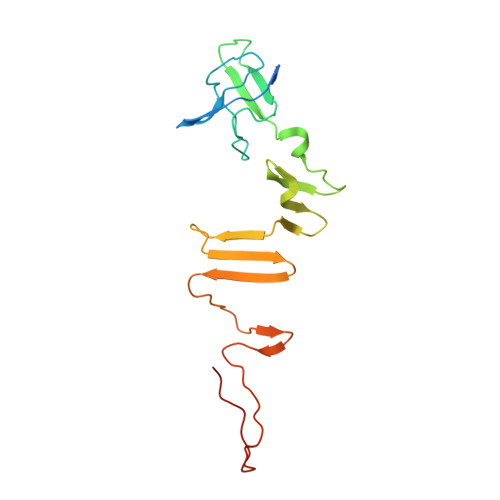
An extensive disulfide bond network prevents tail contraction in Agrobacterium tumefaciens phage Milano.
Sonani, R.R., Palmer, L.K., Esteves, N.C., Horton, A.A., Sebastian, A.L., Kelly, R.J., Wang, F., Kreutzberger, M.A.B., Russell, W.K., Leiman, P.G., Scharf, B.E., Egelman, E.H.(2024) Nat Commun 15: 756-756
- PubMed: 38272938
- DOI: https://doi.org/10.1038/s41467-024-44959-z
- Primary Citation of Related Structures:
8FOP, 8FOU, 8FOY, 8FQC - PubMed Abstract:
A contractile sheath and rigid tube assembly is a widespread apparatus used by bacteriophages, tailocins, and the bacterial type VI secretion system to penetrate cell membranes. In this mechanism, contraction of an external sheath powers the motion of an inner tube through the membrane. The structure, energetics, and mechanism of the machinery imply rigidity and straightness. The contractile tail of Agrobacterium tumefaciens bacteriophage Milano is flexible and bent to varying degrees, which sets it apart from other contractile tail-like systems. Here, we report structures of the Milano tail including the sheath-tube complex, baseplate, and putative receptor-binding proteins. The flexible-to-rigid transformation of the Milano tail upon contraction can be explained by unique electrostatic properties of the tail tube and sheath. All components of the Milano tail, including sheath subunits, are crosslinked by disulfides, some of which must be reduced for contraction to occur. The putative receptor-binding complex of Milano contains a tailspike, a tail fiber, and at least two small proteins that form a garland around the distal ends of the tailspikes and tail fibers. Despite being flagellotropic, Milano lacks thread-like tail filaments that can wrap around the flagellum, and is thus likely to employ a different binding mechanism.
- Department of Biochemistry and Molecular Genetics, University of Virginia School of Medicine, Charlottesville, VA, 22903, USA.
Organizational Affiliation: