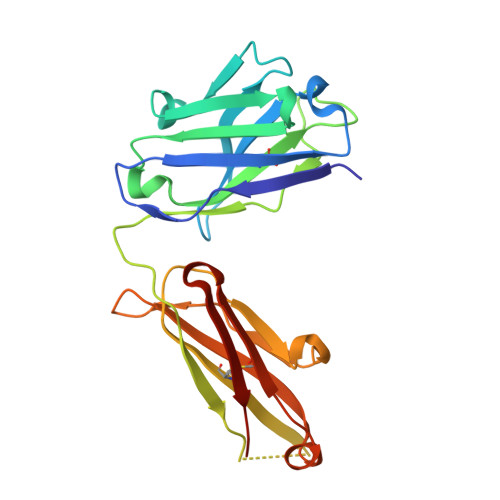
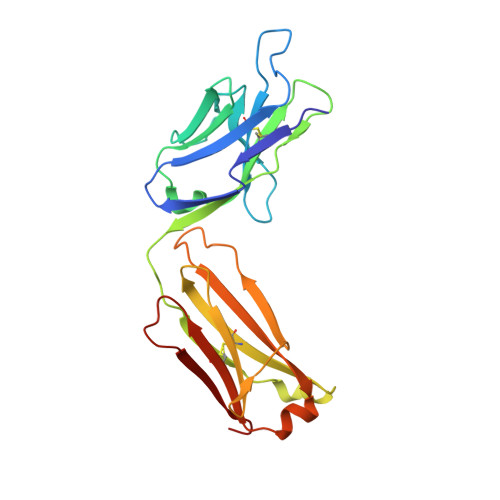
Molecular and functional properties of human Plasmodium falciparum CSP C-terminus antibodies.
Oludada, O.E., Costa, G., Burn Aschner, C., Obraztsova, A.S., Prieto, K., Canetta, C., Hoffman, S.L., Kremsner, P.G., Mordmuller, B., Murugan, R., Julien, J.P., Levashina, E.A., Wardemann, H.(2023) EMBO Mol Med 15: e17454-e17454
- PubMed: 37082831
- DOI: https://doi.org/10.15252/emmm.202317454
- Primary Citation of Related Structures:
8FG0 - PubMed Abstract:
Human monoclonal antibodies (mAbs) against the central repeat and junction domain of Plasmodium falciparum circumsporozoite protein (PfCSP) have been studied extensively to guide malaria vaccine design compared to antibodies against the PfCSP C terminus. Here, we describe the molecular characteristics and protective potential of 73 germline and mutated human mAbs against the highly immunogenic PfCSP C-terminal domain. Two mAbs recognized linear epitopes in the C-terminal linker with sequence similarity to repeat and junction motifs, whereas all others targeted conformational epitopes in the α-thrombospondin repeat (α-TSR) domain. Specificity for the polymorphic Th2R/Th3R but not the conserved RII+/CS.T3 region in the α-TSR was associated with IGHV3-21/IGVL3-21 or IGLV3-1 gene usage. Although the C terminus specific mAbs showed signs of more efficient affinity maturation and class-switching compared to anti-repeat mAbs, live sporozoite binding and inhibitory activity was limited to a single C-linker reactive mAb with cross-reactivity to the central repeat and junction. The data provide novel insights in the human anti-C-linker and anti-α-TSR antibody response that support exclusion of the PfCSP C terminus from malaria vaccine designs.
- B Cell Immunology, German Cancer Research Center, Heidelberg, Germany.
Organizational Affiliation: