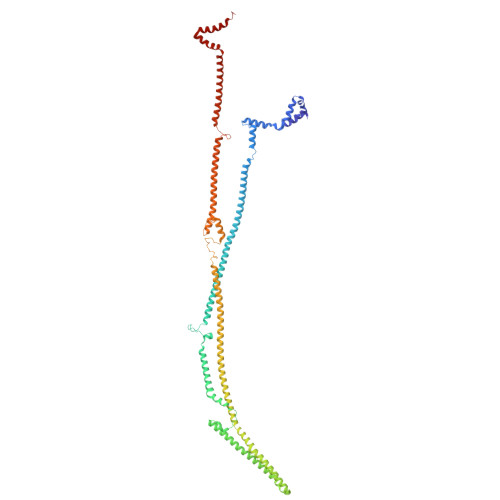
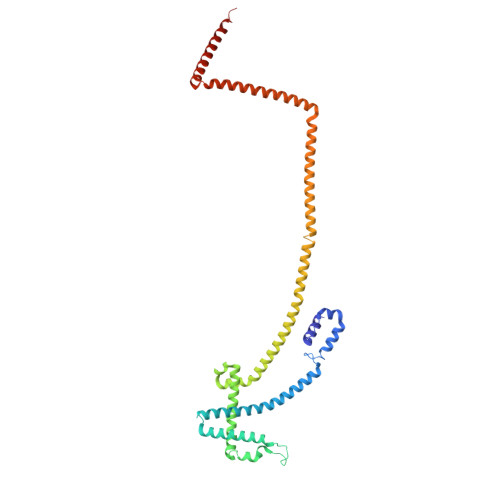
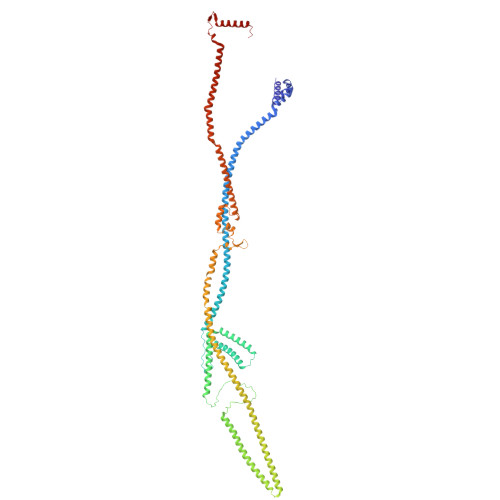
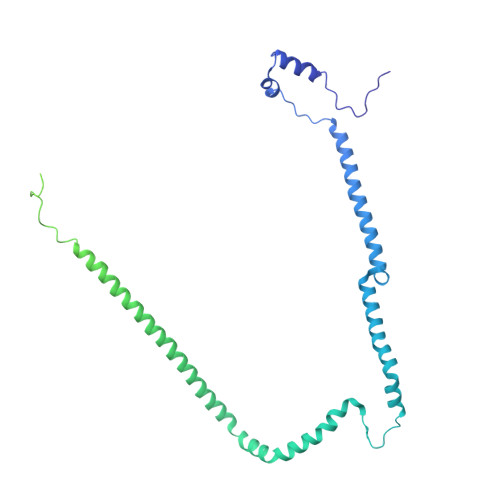
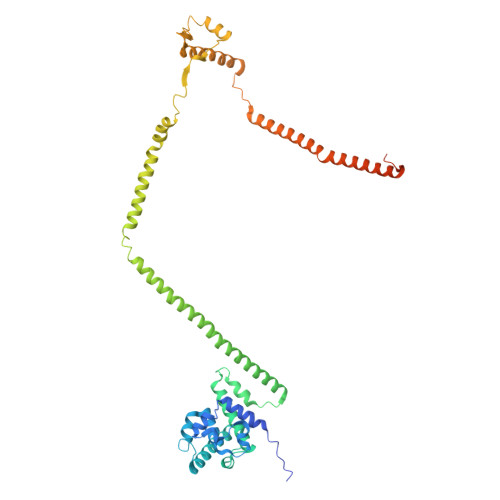
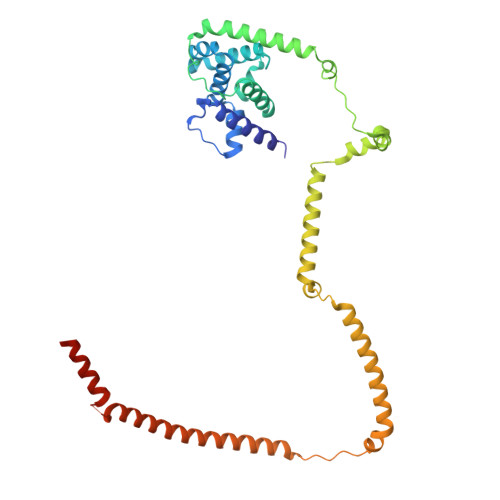
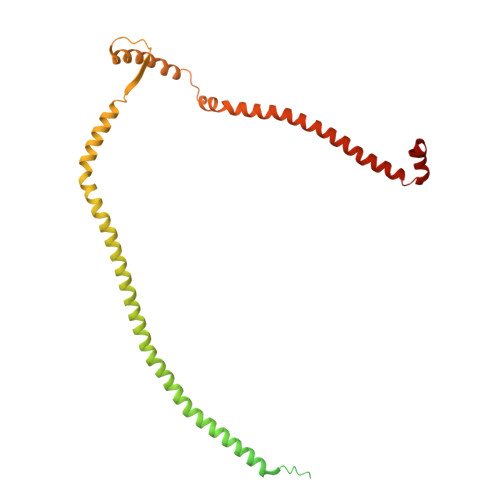
Integrated model of the vertebrate augmin complex.
Travis, S.M., Mahon, B.P., Huang, W., Ma, M., Rale, M.J., Kraus, J., Taylor, D.J., Zhang, R., Petry, S.(2023) Nat Commun 14: 2072-2072
- PubMed: 37055408
- DOI: https://doi.org/10.1038/s41467-023-37519-4
- Primary Citation of Related Structures:
8FCK - PubMed Abstract:
Accurate segregation of chromosomes is required to maintain genome integrity during cell division. This feat is accomplished by the microtubule-based spindle. To build a spindle rapidly and with high fidelity, cells take advantage of branching microtubule nucleation, which rapidly amplifies microtubules during cell division. Branching microtubule nucleation relies on the hetero-octameric augmin complex, but lack of structure information about augmin has hindered understanding how it promotes branching. In this work, we combine cryo-electron microscopy, protein structural prediction, and visualization of fused bulky tags via negative stain electron microscopy to identify the location and orientation of each subunit within the augmin structure. Evolutionary analysis shows that augmin's structure is highly conserved across eukaryotes, and that augmin contains a previously unidentified microtubule binding site. Thus, our findings provide insight into the mechanism of branching microtubule nucleation.
- Department of Molecular Biology, Princeton University, Princeton, NJ, USA.
Organizational Affiliation: