Apo structure of (N1G37) Methyltransferase from Mycobacterium avium.
Isaacson, B., Brylewski, J., Schlegel, F., Cortes, J., Lavallee, T., Mehta, K., Young, A., Doti, L., Battaile, K.P., Stojanoff, V., Perez, A., Bolen, R.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| tRNA (guanine-N(1)-)-methyltransferase | 260 | Mycobacterium avium | Mutation(s): 0 Gene Names: trmD, MAP_2974c EC: 2.1.1.228 | 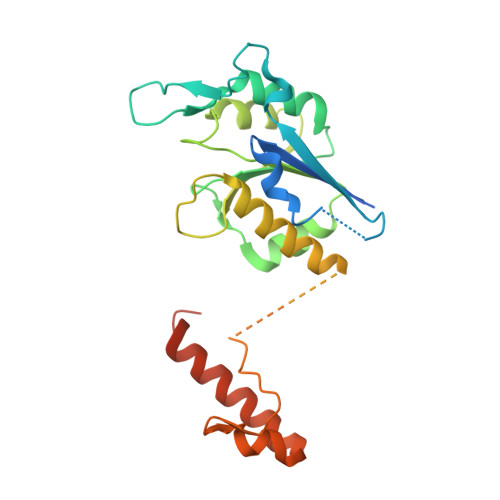 | |
UniProt | |||||
Find proteins for Q73VN9 (Mycolicibacterium paratuberculosis (strain ATCC BAA-968 / K-10)) Explore Q73VN9 Go to UniProtKB: Q73VN9 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q73VN9 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 71.2 | α = 90 |
| b = 71.2 | β = 90 |
| c = 240.405 | γ = 90 |
| Software Name | Purpose |
|---|---|
| BUSTER | refinement |
| autoPROC | data processing |
| XDS | data reduction |
| Aimless | data scaling |
| STARANISO | data scaling |
| EPMR | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Brookhaven National Laboratory (BNL) | United States | DE-SC0012704 |