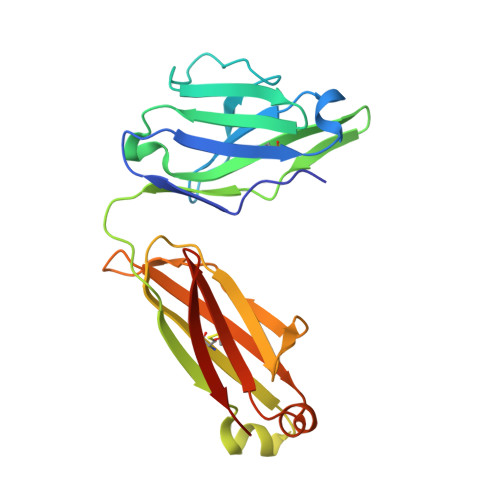
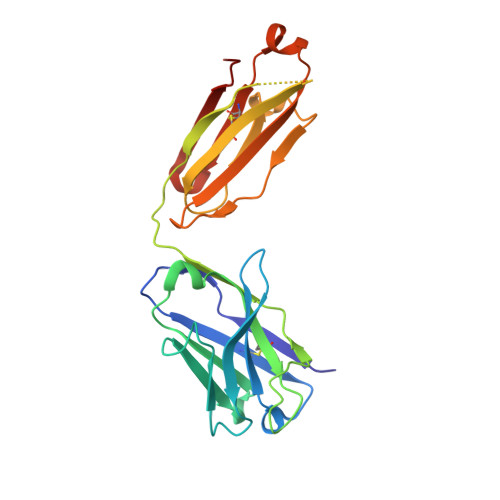
Three-dimensional structure of murine anti-p-azophenylarsonate Fab 36-71. 1. X-ray crystallography, site-directed mutagenesis, and modeling of the complex with hapten.
Strong, R.K., Campbell, R., Rose, D.R., Petsko, G.A., Sharon, J., Margolies, M.N.(1991) Biochemistry 30: 3739-3748
- PubMed: 2015229
- DOI: https://doi.org/10.1021/bi00229a022
- Primary Citation of Related Structures:
6FAB, 8FAB - PubMed Abstract:
The structure of the antigen-binding fragment (Fab) of an anti-p-azophenylarsonate monoclonal antibody, 36-71, bearing a major cross-reactive idiotype of A/J mice has been refined to an R factor of 24.8% at a resolution of 1.85 A. The previously solved partial structure of this Fab at a resolution of 2.9 A (Rose et al., 1990) was used as an initial model for refinement against the high-resolution data. The complex with hapten has been modeled by docking the small-molecule crystal structure of phenylarsonic acid into the structure of the native Fab on the basis of a low-resolution electron density map of the complex. In this model, residue Arg-96 in the light chain and residues Asn-35, Trp-47, and Ser-99 in the heavy chain contact the arsonate moiety of the hapten; an additional bond is found between the arsonate group and a tightly bound water molecule. The phenyl moiety of the hapten packs against two tyrosine side chains at positions 50 and 106 in the heavy chain. Residue Arg-96 in the light chain had been implicated as involved in hapten binding on the basis of previous experiments, and indeed, this residue appears to play a crucial role in this model. Experiments employing site-directed mutagenesis directly support this conclusion. The heavy-chain complementarity-determining regions have novel conformations not previously observed in immunoglobulins except for the recently solved anti-p-azophenylarsonate Fab R 19.9 (Lascombe et al., 1989).
- California Institute of Technology, Pasadena 91125.
Organizational Affiliation: