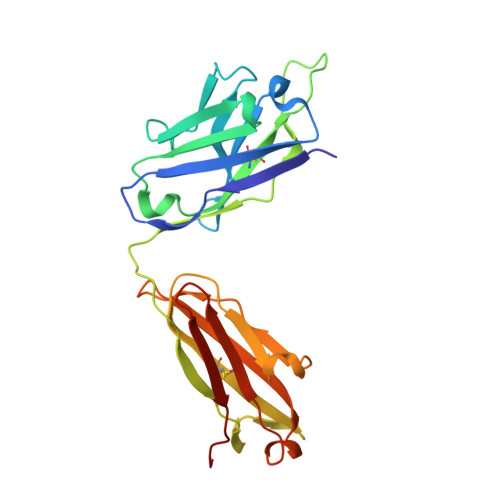
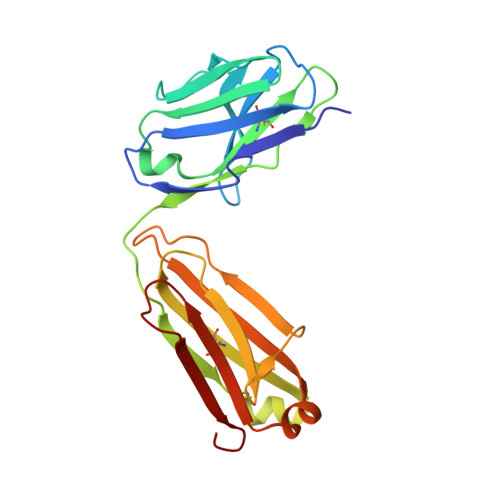
Molecular determinants of cross-reactivity and potency by VH3-33 antibodies against the Plasmodium falciparum circumsporozoite protein.
Thai, E., Murugan, R., Binter, S., Burn Aschner, C., Prieto, K., Kassardjian, A., Obraztsova, A.S., Kang, R.W., Flores-Garcia, Y., Mathis-Torres, S., Li, K., Horn, G.Q., Huntwork, R.H.C., Bolscher, J.M., de Bruijni, M.H.C., Sauerwein, R., Dennison, S.M., Tomaras, G.D., Zavala, F., Kellam, P., Wardemann, H., Julien, J.P.(2023) Cell Rep 42: 113330-113330
- PubMed: 38007690
- DOI: https://doi.org/10.1016/j.celrep.2023.113330
- Primary Citation of Related Structures:
8F95, 8F9E, 8F9F, 8F9S, 8F9T, 8F9U, 8F9V, 8F9W, 8FA6, 8FA7, 8FA8, 8FA9, 8FAN, 8FAS, 8FAT, 8FB5, 8FB6, 8FB7, 8FB8, 8FBA, 8FDC, 8FDD - PubMed Abstract:
IGHV3-33-encoded antibodies are prevalent in the human humoral response against the Plasmodium falciparum circumsporozoite protein (PfCSP). Among VH3-33 antibodies, cross-reactivity between PfCSP major repeat (NANP), minor (NVDP), and junctional (NPDP) motifs is associated with high affinity and potent parasite inhibition. However, the molecular basis of antibody cross-reactivity and the relationship with efficacy remain unresolved. Here, we perform an extensive structure-function characterization of 12 VH3-33 anti-PfCSP monoclonal antibodies (mAbs) with varying degrees of cross-reactivity induced by immunization of mice expressing a human immunoglobulin gene repertoire. We identify residues in the antibody paratope that mediate cross-reactive binding and delineate four distinct epitope conformations induced by antibody binding, with one consistently associated with high protective efficacy and another that confers comparably potent inhibition of parasite liver invasion. Our data show a link between molecular features of cross-reactive VH3-33 mAb binding to PfCSP and mAb potency, relevant for the development of antibody-based interventions against malaria.
- Program in Molecular Medicine, The Hospital for Sick Children Research Institute, Toronto, ON M5G 0A4, Canada; Department of Biochemistry, University of Toronto, Toronto, ON M5S 1A8, Canada.
Organizational Affiliation: