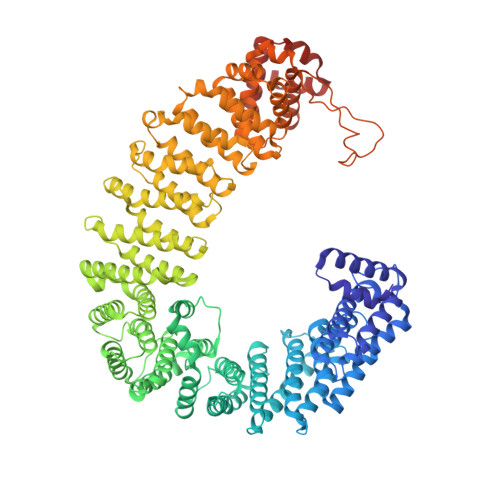
Mechanism of RanGTP priming H2A-H2B release from Kap114 in an atypical RanGTP•Kap114•H2A-H2B complex.
Jiou, J., Shaffer, J.M., Bernades, N.E., Fung, H.Y.J., Kikumoto Dias, J., D'Arcy, S., Chook, Y.M.(2023) Proc Natl Acad Sci U S A 120: e2301199120-e2301199120
- PubMed: 37450495
- DOI: https://doi.org/10.1073/pnas.2301199120
- Primary Citation of Related Structures:
8F0X, 8F19, 8F1E, 8F7A - PubMed Abstract:
Previously, we showed that the nuclear import receptor Importin-9 wraps around the H2A-H2B core to chaperone and transport it from the cytoplasm to the nucleus. However, unlike most nuclear import systems where RanGTP dissociates cargoes from their importins, RanGTP binds stably to the Importin-9•H2A-H2B complex, and formation of the ternary RanGTP•Importin-9•H2A-H2B complex facilitates H2A-H2B release to the assembling nucleosome. It was unclear how RanGTP and the cargo H2A-H2B can bind simultaneously to an importin, and how interactions of the three components position H2A-H2B for release. Here, we show cryo-EM structures of Importin-9•RanGTP and of its yeast homolog Kap114, including Kap114•RanGTP, Kap114•H2A-H2B, and RanGTP•Kap114•H2A-H2B, to explain how the conserved Kap114 binds H2A-H2B and RanGTP simultaneously and how the GTPase primes histone transfer to the nucleosome. In the ternary complex, RanGTP binds to the N-terminal repeats of Kap114 in the same manner as in the Kap114/Importin-9•RanGTP complex, and H2A-H2B binds via its acidic patch to the Kap114 C-terminal repeats much like in the Kap114/Importin-9•H2A-H2B complex. Ran binds to a different conformation of Kap114 in the ternary RanGTP•Kap114•H2A-H2B complex. Here, Kap114 no longer contacts the H2A-H2B surface proximal to the H2A docking domain that drives nucleosome assembly, positioning it for transfer to the assembling nucleosome or to dedicated H2A-H2B chaperones in the nucleus.
- Department of Pharmacology, University of Texas Southwestern Medical Center, Dallas, TX 75390.
Organizational Affiliation: