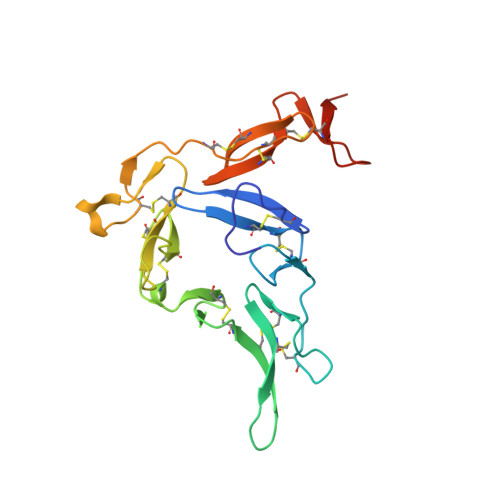
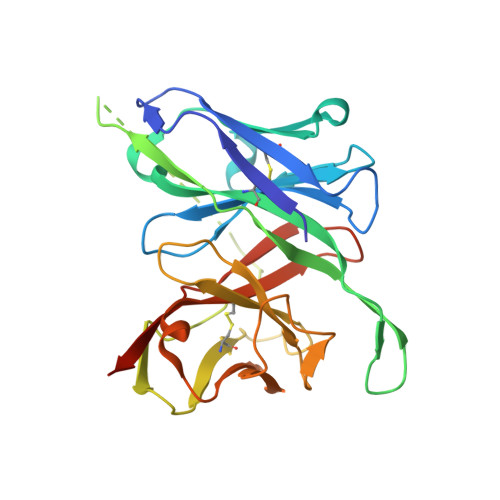
A human antibody epitope map of the malaria vaccine antigen Pfs25.
Shukla, N., Tang, W.K., Coelho, C.H., Long, C.A., Healy, S.A., Sagara, I., Miura, K., Duffy, P.E., Tolia, N.H.(2023) NPJ Vaccines 8: 108-108
- PubMed: 37542029
- DOI: https://doi.org/10.1038/s41541-023-00712-z
- Primary Citation of Related Structures:
8EZK, 8EZL, 8EZM - PubMed Abstract:
Pfs25 is a leading antigen for a malaria transmission-blocking vaccine and shows moderate transmission-blocking activity and induction of rapidly decreasing antibody titers in clinical trials. A comprehensive definition of all transmission-reducing epitopes of Pfs25 will inform structure-guided design to enhance Pfs25-based vaccines, leading to potent transmission-blocking activity. Here, we compiled a detailed human antibody epitope map comprising epitope binning data and structures of multiple human monoclonal antibodies, including three new crystal structures of Pfs25 in complex with transmission-reducing antibodies from Malian volunteers immunized with Pfs25 conjugated to EPA and adjuvanted with AS01. These structures revealed additional epitopes in Pfs25 capable of reducing transmission and expanded this characterization to malaria-exposed humans. This work informs immunogen design to focus the antibody response to transmission-reducing epitopes of Pfs25, enabling development of more potent transmission-blocking vaccines for malaria.
- Host-Pathogen Interactions and Structural Vaccinology Section, Laboratory of Malaria Immunology and Vaccinology, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, USA.
Organizational Affiliation: