Structural and dynamical basis for the interaction of HSP70-EEVD with JDP Sis1
Matos, C.O., Pinheiro, G.M.S., Ramos, C.H.I., Almeida, F.C.L.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Protein SIS1 | 84 | Saccharomyces cerevisiae S288C | Mutation(s): 0 Gene Names: SIS1, YNL007C, N2879 | 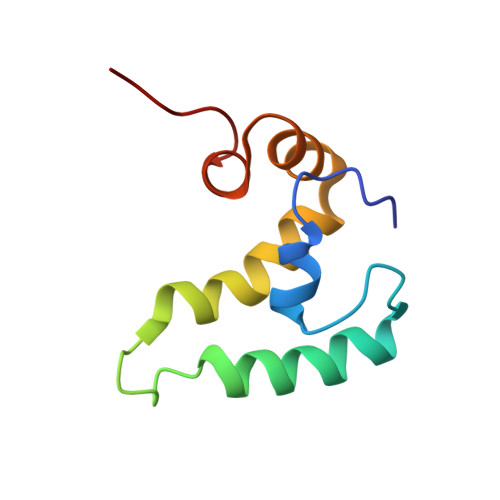 | |
UniProt | |||||
Find proteins for P25294 (Saccharomyces cerevisiae (strain ATCC 204508 / S288c)) Explore P25294 Go to UniProtKB: P25294 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P25294 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Funding Organization | Location | Grant Number |
|---|---|---|
| Sao Paulo Research Foundation (FAPESP) | Brazil | 2019/16114-1 |
| Sao Paulo Research Foundation (FAPESP) | Brazil | 2017/26131-5 |
| Brazilian National Council for Scientific and Technological Development (CNPq) | Brazil | 313517/2021-5 |