Evaluation of novel pyrazol-4-yl pyridine derivatives possessing arylsulfonamide tethers as c-Jun N-terminal kinase (JNK) inhibitors in leukemia cells.
Mersal, K.I., Abdel-Maksoud, M.S., Ali, E.M.H., Ammar, U.M., Zaraei, S.O., Haque, M.M., Das, T., Hassan, N.F., Kim, E.E., Lee, J.S., Park, H., Lee, K.H., El-Gamal, M.I., Kim, H.K., Ibrahim, T.M., Oh, C.H.(2023) Eur J Med Chem 261: 115779-115779
- PubMed: 37776574
- DOI: https://doi.org/10.1016/j.ejmech.2023.115779
- Primary Citation of Related Structures:
8ENJ - PubMed Abstract:
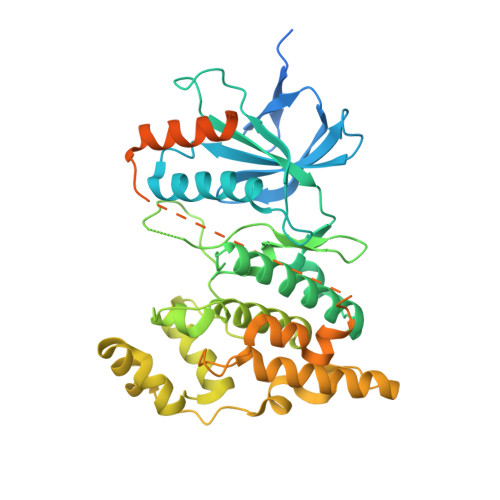
A series of 36 pyrazol-4-yl pyridine derivatives (8a-i, 9a-i, 10a-i, and 11a-i) was designed, synthesized, and evaluated for its antiproliferative activity over NCI-60 cancer cell line panel and inhibitory effect against JNK isoforms (JNK1, JNK2, and JNK3). All the synthesized compounds were tested against the NCI-60 cancer cell line panel. Compounds 11b, 11c, 11g, and 11i were selected to determine their GI 50s and exerted a superior potency over the reference standard SP600125 against the tested cell lines. 11c showed a GI 50 of 1.28 μM against K562 leukemic cells. Vero cells were used to assess 11c cytotoxicity compared to the tested cancer cells. The target compounds were tested against hJNK isoforms in which compound 11e exhibited the highest potency against JNK isoforms with IC 50 values of 1.81, 12.7, and 10.5 nM against JNK1, JNK2, and JNK3, respectively. Kinase profiling of 11e showed higher JNK selectivity in 50 kinase panels. Compounds 11c and 11e showed cell population arrest at the G2/M phase, induced early apoptosis, and slightly inhibited beclin-1 production at higher concentrations in K562 leukemia cells relative to SP600125. NanoBRET assay of 11e showed intracellular JNK1 inhibition with an IC 50 of 2.81 μM. Also, it inhibited CYP2D6 and 3A4 with different extent and its hERG activity showed little cardiac toxicity with an IC 50 of 4.82 μM. hJNK3 was used as a template to generate the hJNK1 crystal structure to explore the binding mode of 11e (PDB ID: 8ENJ) with a resolution of 2.8 °A and showed a typical type I kinase inhibition against hJNK1. Binding energy scores showed that selectivity of 11e towards JNK1 could be attributed to additional hydrophobic interactions relative to JNK3.
- Pharmaceutical Chemistry Department, Faculty of Pharmacy, Modern University for Technology and Information (MTI), Cairo, 12055, Egypt; University of Science & Technology (UST), Daejeon, Yuseong-gu, 34113, Republic of Korea; Center of Biomaterials, Korea Institute of Science & Technology (KIST School), Seoul, Seongbuk-gu, 02792, Republic of Korea.
Organizational Affiliation: