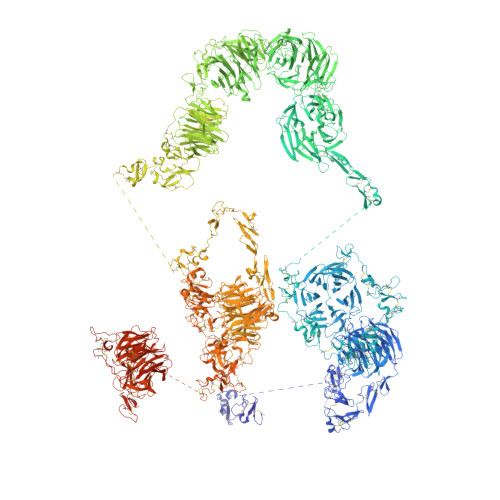
Structures of LRP2 reveal a molecular machine for endocytosis.
Beenken, A., Cerutti, G., Brasch, J., Guo, Y., Sheng, Z., Erdjument-Bromage, H., Aziz, Z., Robbins-Juarez, S.Y., Chavez, E.Y., Ahlsen, G., Katsamba, P.S., Neubert, T.A., Fitzpatrick, A.W.P., Barasch, J., Shapiro, L.(2023) Cell 186: 821
- PubMed: 36750096
- DOI: https://doi.org/10.1016/j.cell.2023.01.016
- Primary Citation of Related Structures:
8EM4, 8EM7 - PubMed Abstract:
The low-density lipoprotein (LDL) receptor-related protein 2 (LRP2 or megalin) is representative of the phylogenetically conserved subfamily of giant LDL receptor-related proteins, which function in endocytosis and are implicated in diseases of the kidney and brain. Here, we report high-resolution cryoelectron microscopy structures of LRP2 isolated from mouse kidney, at extracellular and endosomal pH. The structures reveal LRP2 to be a molecular machine that adopts a conformation for ligand binding at the cell surface and for ligand shedding in the endosome. LRP2 forms a homodimer, the conformational transformation of which is governed by pH-sensitive sites at both homodimer and intra-protomer interfaces. A subset of LRP2 deleterious missense variants in humans appears to impair homodimer assembly. These observations lay the foundation for further understanding the function and mechanism of LDL receptors and implicate homodimerization as a conserved feature of the LRP receptor subfamily.
- Division of Nephrology, Department of Medicine, Columbia University Vagelos College of Physicians and Surgeons, New York, NY 10032, USA.
Organizational Affiliation: