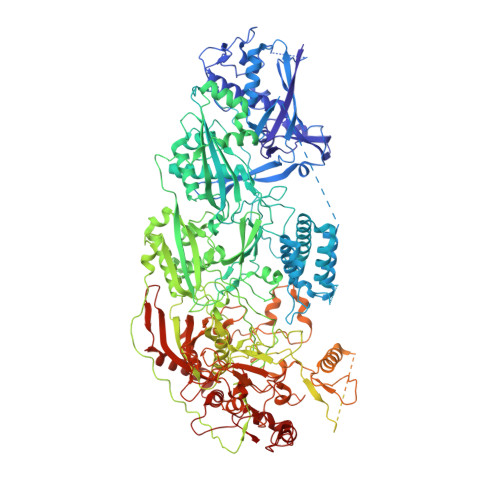
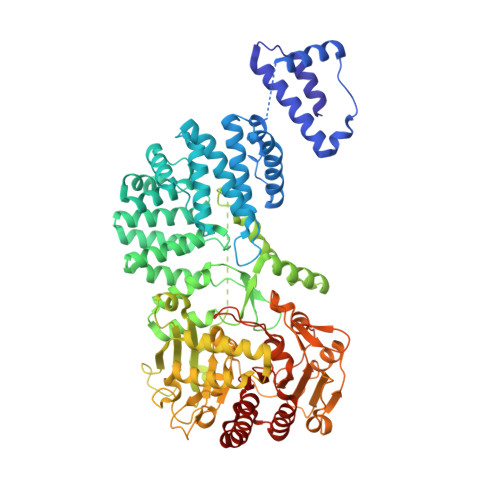
RNA-activated protein cleavage with a CRISPR-associated endopeptidase.
Strecker, J., Demircioglu, F.E., Li, D., Faure, G., Wilkinson, M.E., Gootenberg, J.S., Abudayyeh, O.O., Nishimasu, H., Macrae, R.K., Zhang, F.(2022) Science 378: 874-881
- PubMed: 36423276
- DOI: https://doi.org/10.1126/science.add7450
- Primary Citation of Related Structures:
8EEX, 8EEY - PubMed Abstract:
In prokaryotes, CRISPR-Cas systems provide adaptive immune responses against foreign genetic elements through RNA-guided nuclease activity. Recently, additional genes with non-nuclease functions have been found in genetic association with CRISPR systems, suggesting that there may be other RNA-guided non-nucleolytic enzymes. One such gene from Desulfonema ishimotonii encodes the TPR-CHAT protease Csx29, which is associated with the CRISPR effector Cas7-11. Here, we demonstrate that this CRISPR-associated protease (CASP) exhibits programmable RNA-activated endopeptidase activity against a sigma factor inhibitor to regulate a transcriptional response. Cryo-electron microscopy of an active and substrate-bound CASP complex reveals an allosteric activation mechanism that reorganizes Csx29 catalytic residues upon target RNA binding. This work reveals an RNA-guided function in nature that can be leveraged for RNA-sensing applications in vitro and in human cells.
- Howard Hughes Medical Institute, Massachusetts Institute of Technology, Cambridge, MA 02139, USA.
Organizational Affiliation: