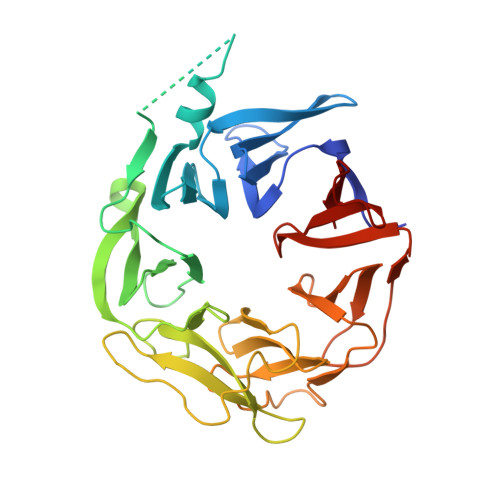
HPK1 citron homology domain regulates phosphorylation of SLP76 and modulates kinase domain interaction dynamics
Chitre, A.S., Wu, P., Walters, B.T., Wang, X., Bouyssou, A., Du, X., Lehoux, I., Fong, R., Arata, A., Chan, J., Wang, D., Franke, Y., Grogan, J.L., Mellman, I., Comps-Agrar, L., Wang, W.(2024) Nat Commun 15: 3725