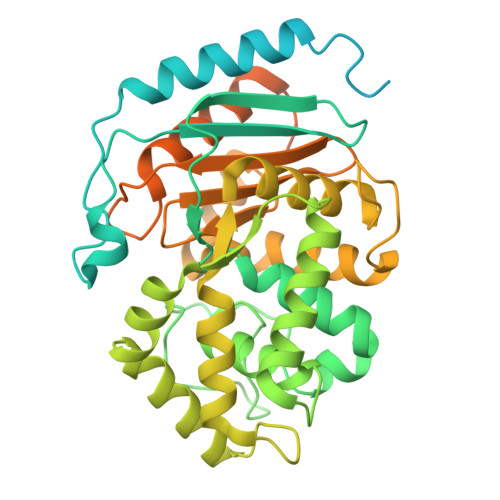
Molecular mechanism of glutaminase activation through filamentation and the role of filaments in mitophagy protection.
Adamoski, D., Dias, M.M., Quesnay, J.E.N., Yang, Z., Zagoriy, I., Steyer, A.M., Rodrigues, C.T., da Silva Bastos, A.C., da Silva, B.N., Costa, R.K.E., de Abreu, F.M.O., Islam, Z., Cassago, A., van Heel, M.G., Consonni, S.R., Mattei, S., Mahamid, J., Portugal, R.V., Ambrosio, A.L.B., Dias, S.M.G.(2023) Nat Struct Mol Biol 30: 1902-1912
- PubMed: 37857822
- DOI: https://doi.org/10.1038/s41594-023-01118-0
- Primary Citation of Related Structures:
8EC6 - PubMed Abstract:
Glutaminase (GLS), which deaminates glutamine to form glutamate, is a mitochondrial tetrameric protein complex. Although inorganic phosphate (Pi) is known to promote GLS filamentation and activation, the molecular basis of this mechanism is unknown. Here we aimed to determine the molecular mechanism of Pi-induced mouse GLS filamentation and its impact on mitochondrial physiology. Single-particle cryogenic electron microscopy revealed an allosteric mechanism in which Pi binding at the tetramer interface and the activation loop is coupled to direct nucleophile activation at the active site. The active conformation is prone to enzyme filamentation. Notably, human GLS filaments form inside tubulated mitochondria following glutamine withdrawal, as shown by in situ cryo-electron tomography of cells thinned by cryo-focused ion beam milling. Mitochondria with GLS filaments exhibit increased protection from mitophagy. We reveal roles of filamentous GLS in mitochondrial morphology and recycling.
- Brazilian Biosciences National Laboratory, Brazilian Center for Research in Energy and Materials, Campinas, Brazil.
Organizational Affiliation: