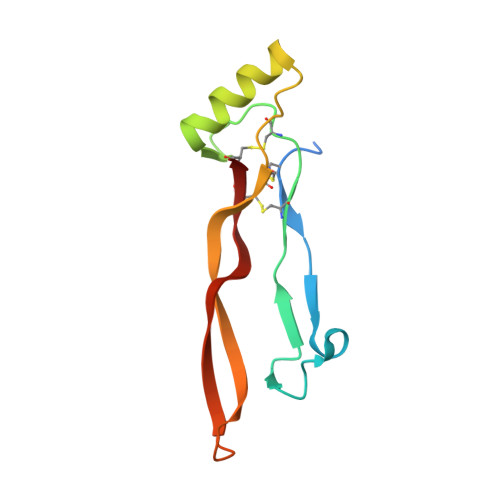
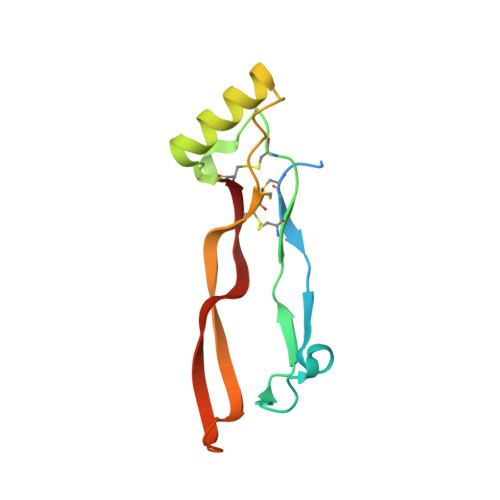
Formation and characterization of BMP2/GDF5 and BMP4/GDF5 heterodimers.
Gipson, G.R., Nolan, K., Kattamuri, C., Kenny, A.P., Agricola, Z., Edwards, N.A., Zinski, J., Czepnik, M., Mullins, M.C., Zorn, A.M., Thompson, T.B.(2023) BMC Biol 21: 16-16
- PubMed: 36726183
- DOI: https://doi.org/10.1186/s12915-023-01522-4
- Primary Citation of Related Structures:
8E3G - PubMed Abstract:
Proteins of the TGFβ family, which are largely studied as homodimers, are also known to form heterodimers with biological activity distinct from their component homodimers. For instance, heterodimers of bone morphogenetic proteins, including BMP2/BMP7, BMP2/BMP6, and BMP9/BMP10, among others, have illustrated the importance of these heterodimeric proteins within the context of TGFβ signaling. In this study, we have determined that mature GDF5 can be combined with mature BMP2 or BMP4 to form BMP2/GDF5 and BMP4/GDF5 heterodimer. Intriguingly, this combination of a BMP2 or BMP4 monomer, which exhibit high affinity to heparan sulfate characteristic to the BMP class, with a GDF5 monomer with low heparan sulfate affinity produces a heterodimer with an intermediate affinity. Using heparin affinity chromatography to purify the heterodimeric proteins, we then determined that both the BMP2/GDF5 and BMP4/GDF5 heterodimers consistently signaled potently across an array of cellular and in vivo systems, while the activities of their homodimeric counterparts were more context dependent. These differences were likely driven by an increase in the combined affinities for the type 1 receptors, Alk3 and Alk6. Furthermore, the X-ray crystal structure of BMP2/GDF5 heterodimer was determined, highlighting the formation of two asymmetric type 1 receptor binding sites that are both unique relative to the homodimers. Ultimately, this method of heterodimer production yielded a signaling molecule with unique properties relative to the homodimeric ligands, including high affinity to multiple type 1 and moderate heparan binding affinity.
- Department of Molecular & Cellular Biosciences, University of Cincinnati College of Medicine, Cincinnati, OH, USA.
Organizational Affiliation: