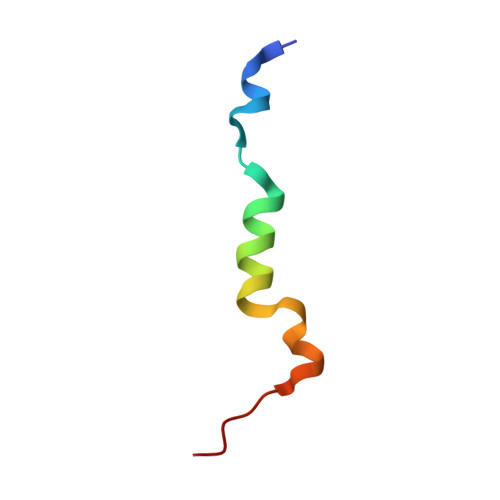
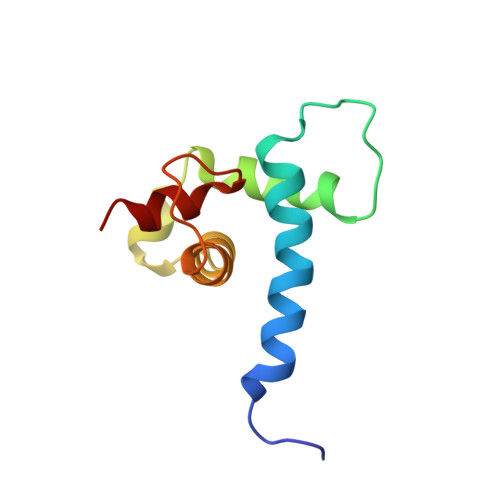
Structural basis of CBP/p300 recruitment by the microphthalmia-associated transcription factor.
Brown, A.D., Vergunst, K.L., Branch, M., Blair, C.M., Dupre, D.J., Baillie, G.S., Langelaan, D.N.(2023) Biochim Biophys Acta Mol Cell Res 1870: 119520-119520
- PubMed: 37353163
- DOI: https://doi.org/10.1016/j.bbamcr.2023.119520
- Primary Citation of Related Structures:
8E1D - PubMed Abstract:
The microphthalmia-associated transcription factor (MITF) is a master regulator of the melanocyte cell lineage. Aberrant MITF activity can lead to multiple malignancies including skin cancer, where it modulates the progression and invasiveness of melanoma. MITF-regulated gene expression requires recruitment of the transcriptional co-regulator CBP/p300, but details of this process are not fully defined. In this study, we investigate the structural and functional interaction between the MITF N-terminal transactivation domain (MITF TAD ) and CBP/p300. Using pulldown assays and nuclear magnetic resonance spectroscopy we determined that MITF TAD is intrinsically disordered and binds to the TAZ1 and TAZ2 domains of CBP/p300 with moderate affinity. The solution-state structure of the MITF TAD :TAZ2 complex reveals that MITF interacts with a hydrophobic surface of TAZ2, while remaining somewhat dynamic. Peptide array and mutagenesis experiments determined that an acidic motif is integral to the MITF TAD :TAZ2 interaction and is necessary for transcriptional activity of MITF. Peptides that bind to the same surface of TAZ2 as MITF TAD , such as the adenoviral protein E1A, are capable of displacing MITF from TAZ2 and inhibiting transactivation. These findings provide insight into co-activator recruitment by MITF that are fundamental to our understanding of MITF targeted gene regulation and melanoma biology.
- Department of Biochemistry & Molecular Biology, Dalhousie University, Halifax, NS B3H 4R2, Canada.
Organizational Affiliation: