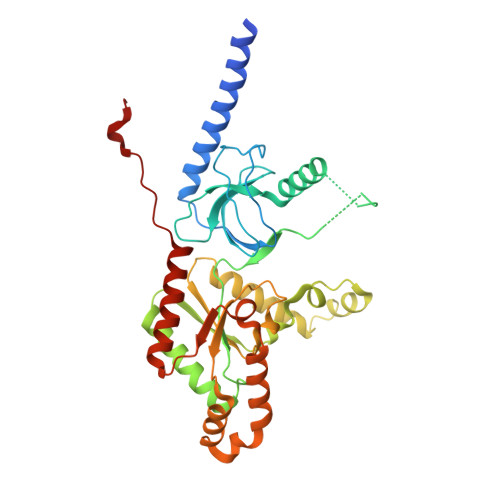
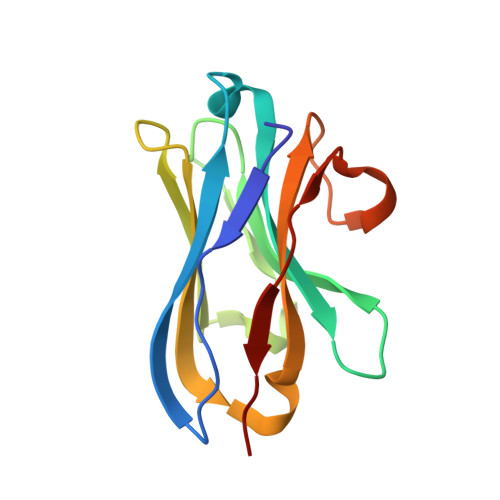
Selective posttranslational inhibition of Ca V beta 1 -associated voltage-dependent calcium channels with a functionalized nanobody.
Morgenstern, T.J., Nirwan, N., Hernandez-Ochoa, E.O., Bibollet, H., Choudhury, P., Laloudakis, Y.D., Ben Johny, M., Bannister, R.A., Schneider, M.F., Minor Jr., D.L., Colecraft, H.M.(2022) Nat Commun 13: 7556-7556
- PubMed: 36494348
- DOI: https://doi.org/10.1038/s41467-022-35025-7
- Primary Citation of Related Structures:
8DAM, 8E0E - PubMed Abstract:
Ca 2+ influx through high-voltage-activated calcium channels (HVACCs) controls diverse cellular functions. A critical feature enabling a singular signal, Ca 2+ influx, to mediate disparate functions is diversity of HVACC pore-forming α 1 and auxiliary Ca V β 1 -Ca V β 4 subunits. Selective Ca V α 1 blockers have enabled deciphering their unique physiological roles. By contrast, the capacity to post-translationally inhibit HVACCs based on Ca V β isoform is non-existent. Conventional gene knockout/shRNA approaches do not adequately address this deficit owing to subunit reshuffling and partially overlapping functions of Ca V β isoforms. Here, we identify a nanobody (nb.E8) that selectively binds Ca V β 1 SH3 domain and inhibits Ca V β 1 -associated HVACCs by reducing channel surface density, decreasing open probability, and speeding inactivation. Functionalizing nb.E8 with Nedd4L HECT domain yielded Chisel-1 which eliminated current through Ca V β 1 -reconstituted Ca V 1/Ca V 2 and native Ca V 1.1 channels in skeletal muscle, strongly suppressed depolarization-evoked Ca 2+ influx and excitation-transcription coupling in hippocampal neurons, but was inert against Ca V β 2 -associated Ca V 1.2 in cardiomyocytes. The results introduce an original method for probing distinctive functions of ion channel auxiliary subunit isoforms, reveal additional dimensions of Ca V β 1 signaling in neurons, and describe a genetically-encoded HVACC inhibitor with unique properties.
- Department of Molecular Pharmacology and Therapeutics, Columbia University Irving Medical Center, New York, NY, USA.
Organizational Affiliation: