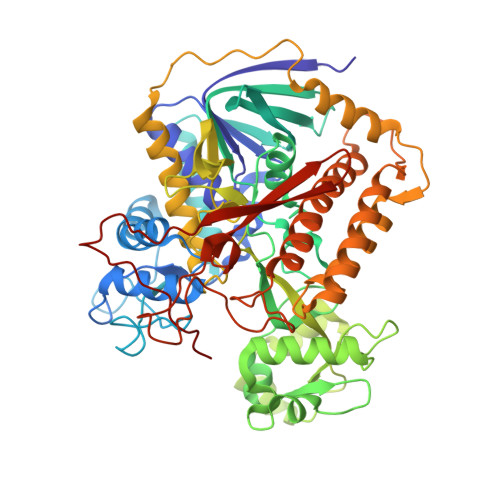
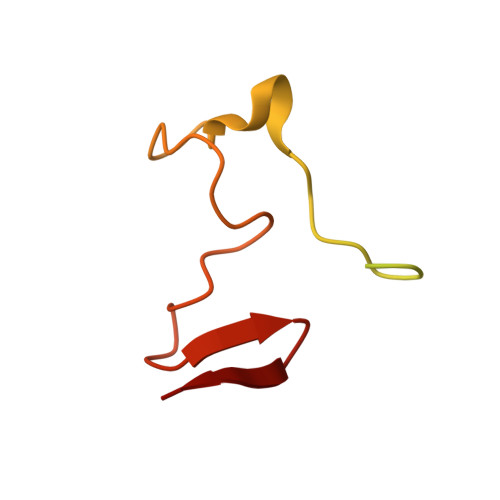
Disordered-to-ordered transitions in assembly factors allow the complex II catalytic subunit to switch binding partners.
Sharma, P., Maklashina, E., Voehler, M., Balintova, S., Dvorakova, S., Kraus, M., Vanova, K.H., Nahacka, Z., Zobalova, R., Boukalova, S., Cunatova, K., Mracek, T., Ghayee, H.K., Pacak, K., Rohlena, J., Neuzil, J., Cecchini, G., Iverson, T.M.(2024) Nat Commun 15: 473-473
- PubMed: 38212624
- DOI: https://doi.org/10.1038/s41467-023-44563-7
- Primary Citation of Related Structures:
8DYD, 8DYE - PubMed Abstract:
Complex II (CII) activity controls phenomena that require crosstalk between metabolism and signaling, including neurodegeneration, cancer metabolism, immune activation, and ischemia-reperfusion injury. CII activity can be regulated at the level of assembly, a process that leverages metastable assembly intermediates. The nature of these intermediates and how CII subunits transfer between metastable complexes remains unclear. In this work, we identify metastable species containing the SDHA subunit and its assembly factors, and we assign a preferred temporal sequence of appearance of these species during CII assembly. Structures of two species show that the assembly factors undergo disordered-to-ordered transitions without the appearance of significant secondary structure. The findings identify that intrinsically disordered regions are critical in regulating CII assembly, an observation that has implications for the control of assembly in other biomolecular complexes.
- Department of Pharmacology, Vanderbilt University, Nashville, TN, 37232, USA.
Organizational Affiliation: