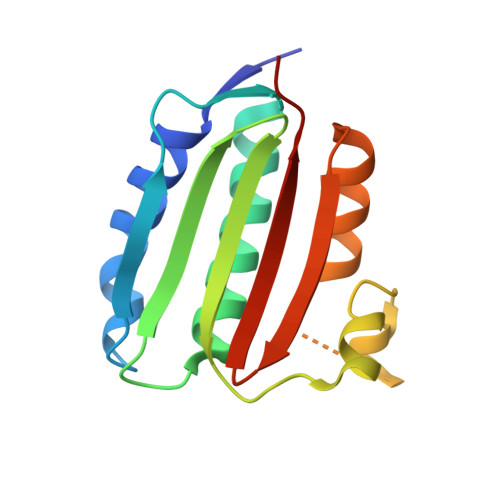
Structure of VanS from vancomycin-resistant enterococci: A sensor kinase with weak ATP binding.
Grasty, K.C., Guzik, C., D'Lauro, E.J., Padrick, S.B., Beld, J., Loll, P.J.(2023) J Biological Chem 299: 103001-103001
- PubMed: 36764524
- DOI: https://doi.org/10.1016/j.jbc.2023.103001
- Primary Citation of Related Structures:
8DVQ, 8DWZ, 8DX0 - PubMed Abstract:
The VanRS two-component system regulates the resistance phenotype of vancomycin-resistant enterococci. VanS is a sensor histidine kinase that responds to the presence of vancomycin by autophosphorylating and subsequently transferring the phosphoryl group to the response regulator, VanR. The phosphotransfer activates VanR as a transcription factor, which initiates the expression of resistance genes. Structural information about VanS proteins has remained elusive, hindering the molecular-level understanding of their function. Here, we present X-ray crystal structures for the catalytic and ATP-binding (CA) domains of two VanS proteins, derived from vancomycin-resistant enterococci types A and C. Both proteins adopt the canonical Bergerat fold that has been observed for CA domains of other prokaryotic histidine kinases. We attempted to determine structures for the nucleotide-bound forms of both proteins; however, despite repeated efforts, these forms could not be crystallized, prompting us to measure the proteins' binding affinities for ATP. Unexpectedly, both CA domains displayed low affinities for the nucleotide, with K D values in the low millimolar range. Since these K D values are comparable to intracellular ATP concentrations, this weak substrate binding could reflect a way of regulating expression of the resistance phenotype.
- Department of Biochemistry and Molecular Biology, Drexel University College of Medicine, Philadelphia, Pennsylvania, USA.
Organizational Affiliation: