Solution Structure of the H3 protein
Kelly, M.J.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| H3 Protein | 167 | Escherichia coli | Mutation(s): 0 | 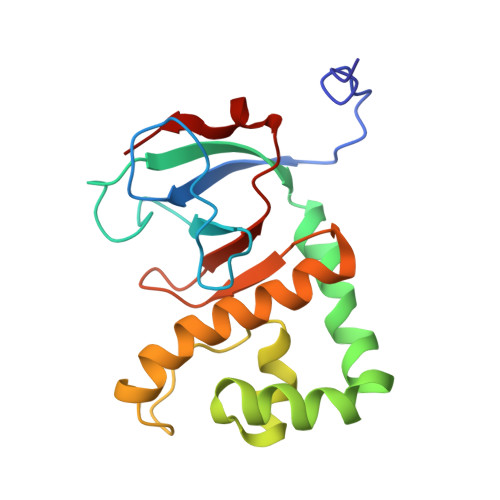 | |
UniProt | |||||
Find proteins for A0AAB0B261 (Escherichia coli) Explore A0AAB0B261 Go to UniProtKB: A0AAB0B261 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0AAB0B261 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Cancer Institute (NIH/NCI) | United States | R01 |