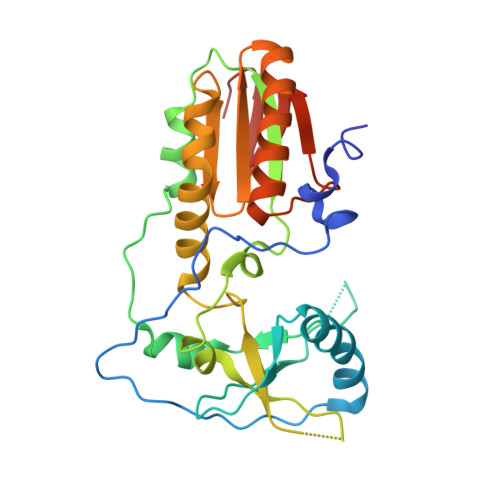
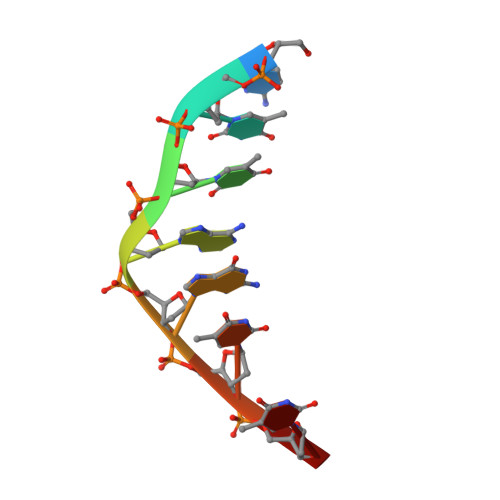
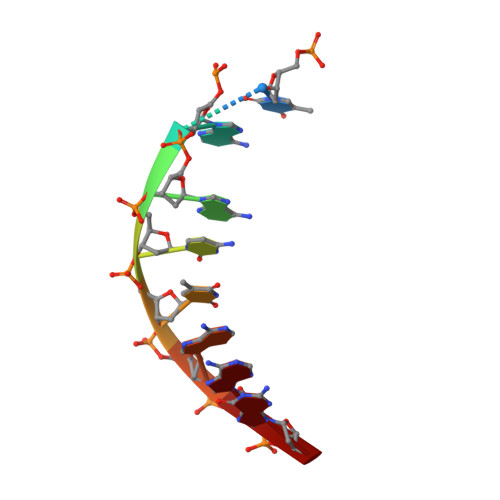
Two distinct rotations of bithiazole DNA intercalation revealed by direct comparison of crystal structures of Co(III)•bleomycin A 2 and B 2 bound to duplex 5'-TAGTT sites.
Goodwin, K.D., Lewis, M.A., Long, E.C., Georgiadis, M.M.(2023) Bioorg Med Chem 77: 117113-117113
- PubMed: 36516684
- DOI: https://doi.org/10.1016/j.bmc.2022.117113
- Primary Citation of Related Structures:
8DW1, 8DW8, 8DWM - PubMed Abstract:
Bleomycins constitute a family of anticancer natural products that bind DNA through intercalation of a C-terminal tail/bithiazole moiety and hydrogen-bonding interactions between the remainder of the drug and the minor groove. The clinical utility of the bleomycins is believed to result from single- and double-strand DNA cleavage mediated by the HOO-Fe(III) form of the drug. The bleomycins also serve as a model system to understand the nature of complex drug-DNA interactions that may guide future DNA-targeted drug discovery. In this study, the impact of the C-terminal tail on bleomycin-DNA interactions was investigated. Toward this goal, we determined two crystal structures of HOO-Co(III)•BLMA 2 "green" (a stable structural analogue of the active HOO-Fe(III) drug) bound to duplex DNA containing 5'-TAGTT, one in which the entire drug is bound (fully bound) and a second with only the C-terminal tail/bithiazole bound (partially bound). The structures reported here were captured by soaking HOO-Co(III)•BLMA 2 into preformed host-guest crystals including a preferred DNA-binding site. While the overall structure of DNA-bound BLMA 2 was found to be similar to those reported earlier at the same DNA site for BLMB 2 , the intercalated bithiazole of BLMB 2 is "flipped" 180˚ relative to DNA-bound BLMA 2 . This finding highlights an unidentified role for the C-terminal tail in directing the intercalation of the bithiazole. In addition, these analyses identified specific bond rotations within the C-terminal domain of the drug that may be relevant for its reorganization and ability to carry out a double-strand DNA cleavage event.
- Department of Biochemistry & Molecular Biology, Indiana University School of Medicine, Indianapolis, IN 46202, USA.
Organizational Affiliation: