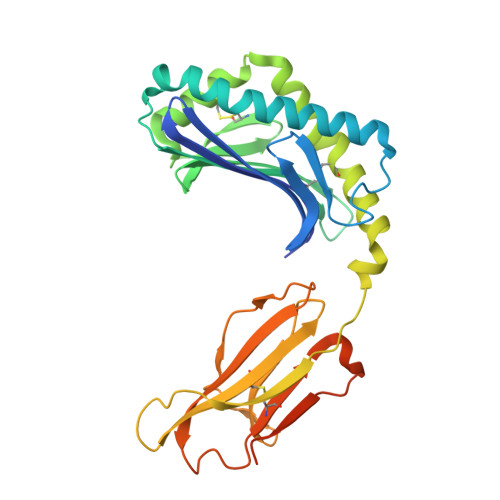
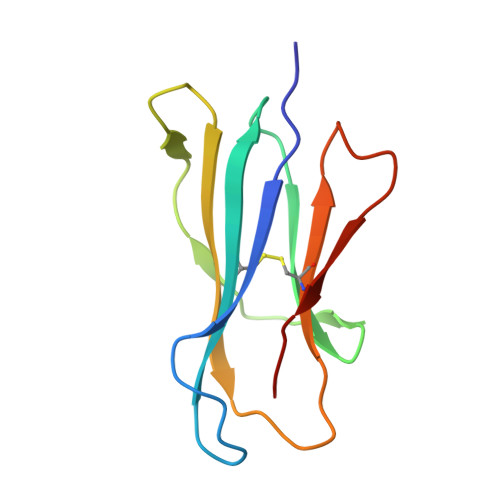
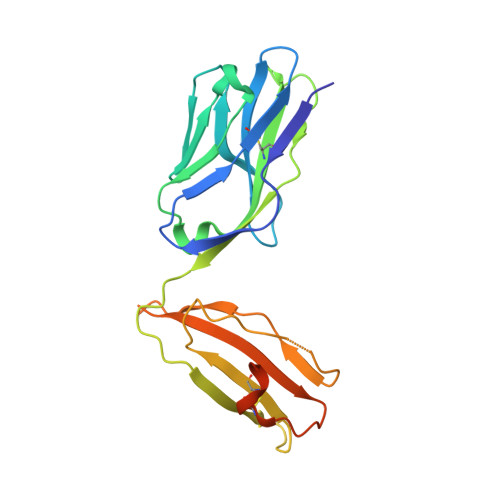
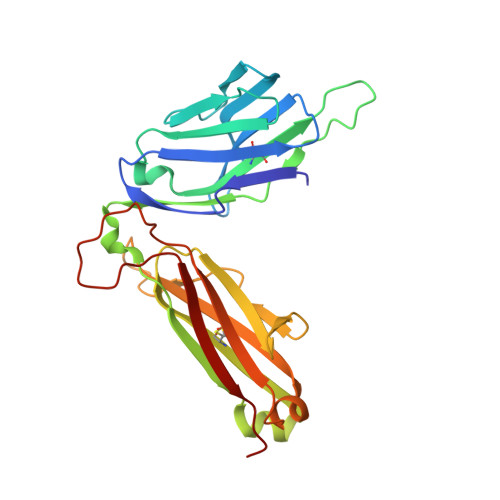
alpha beta T-cell receptor recognition of self-phosphatidylinositol presented by CD1b.
Farquhar, R., Van Rhijn, I., Moody, D.B., Rossjohn, J., Shahine, A.(2023) J Biological Chem 299: 102849-102849
- PubMed: 36587766
- DOI: https://doi.org/10.1016/j.jbc.2022.102849
- Primary Citation of Related Structures:
8DV3, 8DV4 - PubMed Abstract:
CD1 glycoproteins present lipid-based antigens to T-cell receptors (TCRs). A role for CD1b in T-cell-mediated autoreactivity was proposed when it was established that CD1b can present self-phospholipids with short alkyl chains (∼C34) to T cells; however, the structural characteristics of this presentation and recognition are unclear. Here, we report the 1.9 Å resolution binary crystal structure of CD1b presenting a self-phosphatidylinositol-C34:1 and an endogenous scaffold lipid. Moreover, we also determined the 2.4 Å structure of CD1b-phosphatidylinositol complexed to an autoreactive αβ TCR, BC8B. We show that the TCR docks above CD1b and directly contacts the presented antigen, selecting for both the phosphoinositol headgroup and glycerol neck region via antigen remodeling within CD1b and allowing lateral escape of the inositol moiety through a channel formed by the TCR α-chain. Furthermore, through alanine scanning mutagenesis and surface plasmon resonance, we identified key CD1b residues mediating this interaction, with Glu-80 abolishing TCR binding. We in addition define a role for both CD1b α1 and CD1b α2 molecular domains in modulating this interaction. These findings suggest that the BC8B TCR contacts both the presented phospholipid and the endogenous scaffold lipid via a dual mechanism of corecognition. Taken together, these data expand our understanding into the molecular mechanisms of CD1b-mediated T-cell autoreactivity.
- Infection and Immunity Program and Department of Biochemistry and Molecular Biology, Biomedicine Discovery Institute, Monash University, Clayton, Victoria, Australia.
Organizational Affiliation: