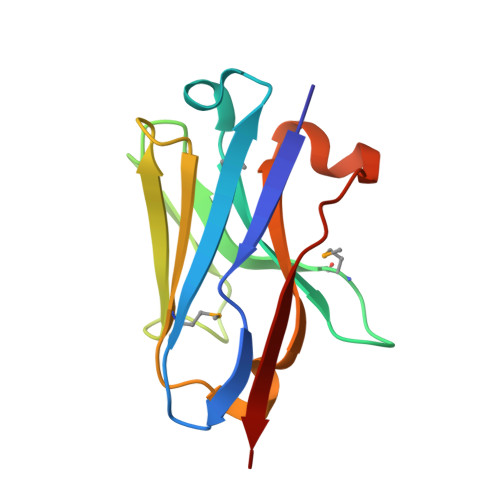
Evolution of nanobodies specific for BCL11A.
Yin, M., Izadi, M., Tenglin, K., Viennet, T., Zhai, L., Zheng, G., Arthanari, H., Dassama, L.M.K., Orkin, S.H.(2023) Proc Natl Acad Sci U S A 120: e2218959120-e2218959120
- PubMed: 36626555
- DOI: https://doi.org/10.1073/pnas.2218959120
- Primary Citation of Related Structures:
8DTN, 8DTU - PubMed Abstract:
Transcription factors (TFs) control numerous genes that are directly relevant to many human disorders. However, developing specific reagents targeting TFs within intact cells is challenging due to the presence of highly disordered regions within these proteins. Intracellular antibodies offer opportunities to probe protein function and validate therapeutic targets. Here, we describe the optimization of nanobodies specific for BCL11A, a validated target for the treatment of hemoglobin disorders. We obtained first-generation nanobodies directed to a region of BCL11A comprising zinc fingers 4 to 6 (ZF456) from a synthetic yeast surface display library, and employed error-prone mutagenesis, structural determination, and molecular modeling to enhance binding affinity. Engineered nanobodies recognized ZF6 and mediated targeted protein degradation (TPD) of BCL11A protein in erythroid cells, leading to the anticipated reactivation of fetal hemoglobin (HbF) expression. Evolved nanobodies distinguished BCL11A from its close paralog BCL11B, which shares an identical DNA-binding specificity. Given the ease of manipulation of nanobodies and their exquisite specificity, nanobody-mediated TPD of TFs should be suitable for dissecting regulatory relationships of TFs and gene targets and validating therapeutic potential of proteins of interest.
- Dana Farber Boston Children's Cancer and Blood Disorders Center, Harvard Medical School, Boston, MA 02115.
Organizational Affiliation: