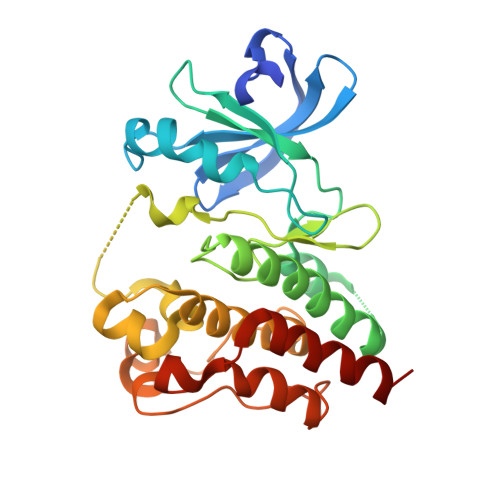
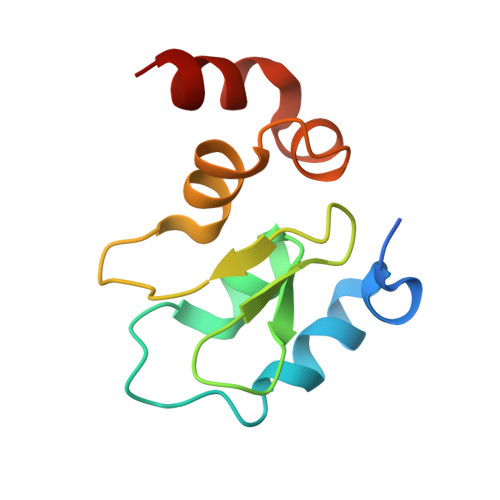
A covalent BTK ternary complex compatible with targeted protein degradation.
Schiemer, J., Maxwell, A., Horst, R., Liu, S., Uccello, D.P., Borzilleri, K., Rajamohan, N., Brown, M.F., Calabrese, M.F.(2023) Nat Commun 14: 1189-1189
- PubMed: 36864023
- DOI: https://doi.org/10.1038/s41467-023-36738-z
- Primary Citation of Related Structures:
8DSF, 8DSO - PubMed Abstract:
Targeted protein degradation using heterobifunctional chimeras holds the potential to expand target space and grow the druggable proteome. Most acutely, this provides an opportunity to target proteins that lack enzymatic activity or have otherwise proven intractable to small molecule inhibition. Limiting this potential, however, is the remaining need to develop a ligand for the target of interest. While a number of challenging proteins have been successfully targeted by covalent ligands, unless this modification affects form or function, it may lack the ability to drive a biological response. Bridging covalent ligand discovery with chimeric degrader design has emerged as a potential mechanism to advance both fields. In this work, we employ a set of biochemical and cellular tools to deconvolute the role of covalent modification in targeted protein degradation using Bruton's tyrosine kinase. Our results reveal that covalent target modification is fundamentally compatible with the protein degrader mechanism of action.
- Discovery Sciences, Pfizer Worldwide Research and Development, Groton, CT, USA.
Organizational Affiliation: