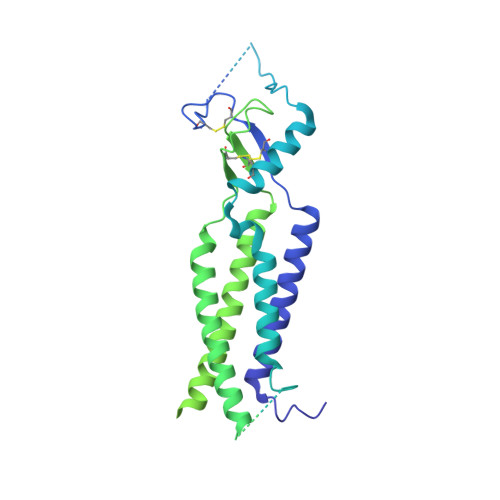
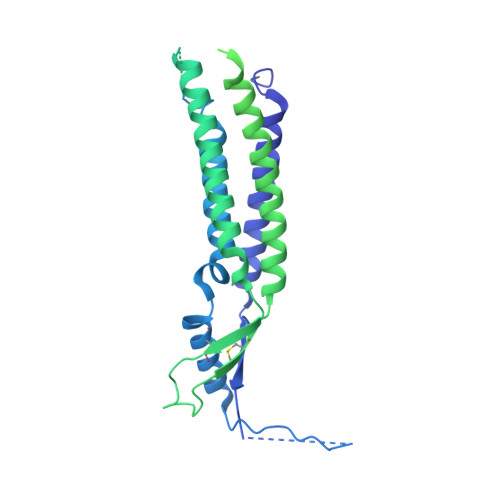
Structural basis for assembly and lipid-mediated gating of LRRC8A:C volume-regulated anion channels.
Kern, D.M., Bleier, J., Mukherjee, S., Hill, J.M., Kossiakoff, A.A., Isacoff, E.Y., Brohawn, S.G.(2023) Nat Struct Mol Biol 30: 841-852
- PubMed: 36928458
- DOI: https://doi.org/10.1038/s41594-023-00944-6
- Primary Citation of Related Structures:
8DR8, 8DRA, 8DRE, 8DRK, 8DRN, 8DRO, 8DRQ, 8DS3, 8DS9, 8DSA, 8F74, 8F75, 8F77, 8F79, 8F7B, 8F7D, 8F7E, 8F7J - PubMed Abstract:
Leucine-rich repeat-containing protein 8 (LRRC8) family members form volume-regulated anion channels activated by hypoosmotic cell swelling. LRRC8 channels are ubiquitously expressed in vertebrate cells as heteromeric assemblies of LRRC8A (SWELL1) and LRRC8B-E subunits. Channels of different subunit composition have distinct properties that explain the functional diversity of LRRC8 currents across cell types. However, the basis for heteromeric LRRC8 channel assembly and function is unknown. Here we leverage a fiducial-tagging strategy to determine single-particle cryo-EM structures of heterohexameric LRRC8A:C channels in multiple conformations. Compared to homomers, LRRC8A:C channels show pronounced differences in architecture due to heterotypic LRR interactions that displace subunits away from the conduction axis and poise the channel for activation. Structures and functional studies further reveal that lipids embedded in the channel pore block ion conduction in the closed state. These results provide insight into determinants for heteromeric LRRC8 channel assembly, activity and gating by lipids.
- Department of Molecular & Cell Biology, University of California, Berkeley, CA, USA.
Organizational Affiliation: