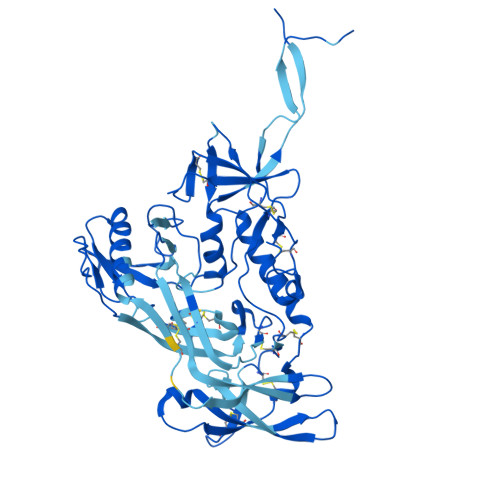
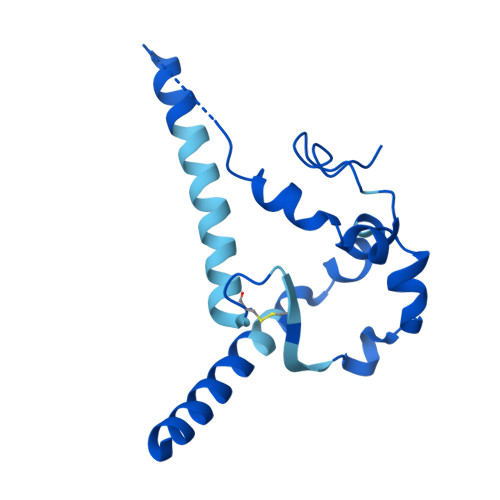
Shared recognition mechanism for HIV-1 envelope-reactive V3 glycan broadly neutralizing B cell lineage maturation in humans and macaques
Gobeil, S.M., Janowska, K., Stalls, V., Mansouri, K., Henderson, R.C., Edwards, R.J., Wiehe, K., Saunders, K., Acharya, P., Williams, W., Haynes, B.F.To be published.