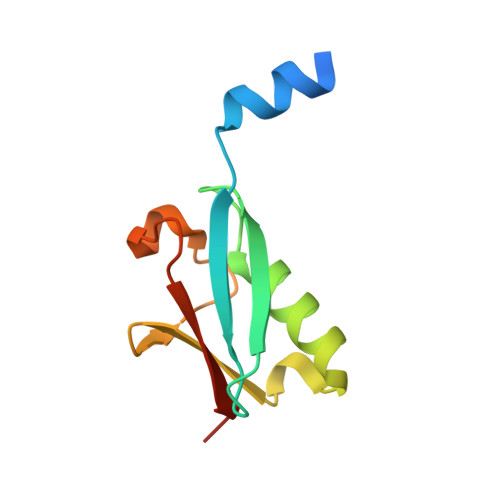
Zinc controls PML nuclear body formation through regulation of a paralog specific auto-inhibition in SUMO1.
Lussier-Price, M., Wahba, H.M., Mascle, X.H., Cappadocia, L., Bourdeau, V., Gagnon, C., Igelmann, S., Sakaguchi, K., Ferbeyre, G., Omichinski, J.G.(2022) Nucleic Acids Res 50: 8331-8348
- PubMed: 35871297
- DOI: https://doi.org/10.1093/nar/gkac620
- Primary Citation of Related Structures:
8DJH, 8DJI - PubMed Abstract:
SUMO proteins are important regulators of many key cellular functions in part through their ability to form interactions with other proteins containing SUMO interacting motifs (SIMs). One characteristic feature of all SUMO proteins is the presence of a highly divergent intrinsically disordered region at their N-terminus. In this study, we examine the role of this N-terminal region of SUMO proteins in SUMO-SIM interactions required for the formation of nuclear bodies by the promyelocytic leukemia (PML) protein (PML-NBs). We demonstrate that the N-terminal region of SUMO1 functions in a paralog specific manner as an auto-inhibition domain by blocking its binding to the phosphorylated SIMs of PML and Daxx. Interestingly, we find that this auto-inhibition in SUMO1 is relieved by zinc, and structurally show that zinc stabilizes the complex between SUMO1 and a phospho-mimetic form of the SIM of PML. In addition, we demonstrate that increasing cellular zinc levels enhances PML-NB formation in senescent cells. Taken together, these results provide important insights into a paralog specific function of SUMO1, and suggest that zinc levels could play a crucial role in regulating SUMO1-SIM interactions required for PML-NB formation and function.
- Département de Biochimie et Médicine Moléculaire, Université de Montréal, Montréal, QC, Canada.
Organizational Affiliation: