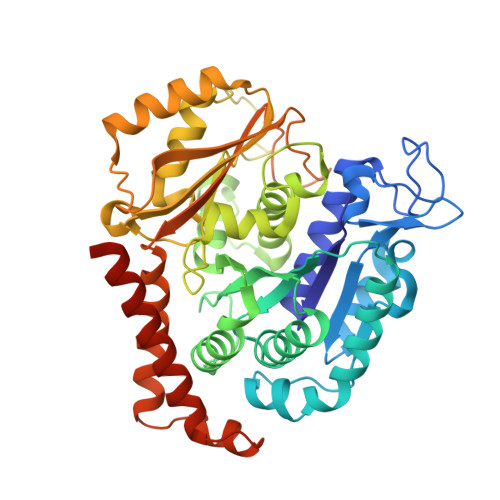
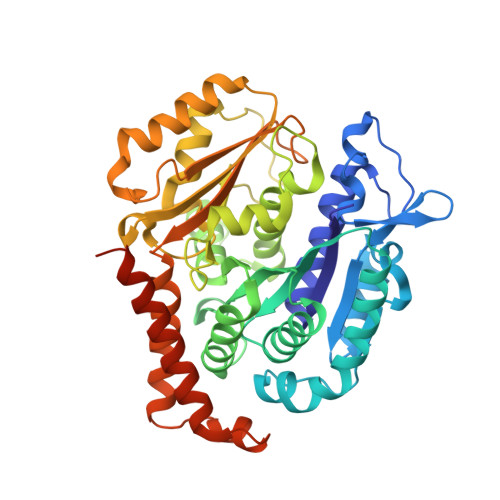
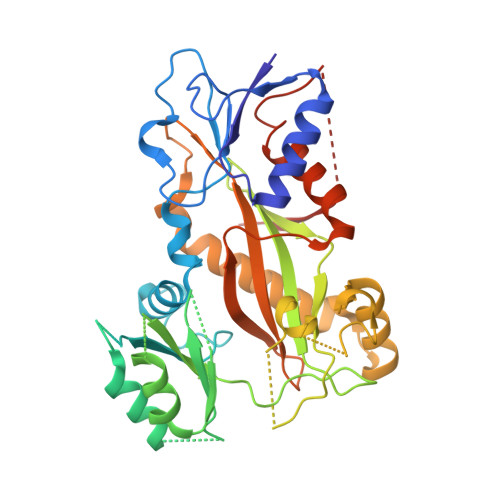
SB226, an inhibitor of tubulin polymerization, inhibits paclitaxel-resistant melanoma growth and spontaneous metastasis.
Deng, S., Banerjee, S., Chen, H., Pochampally, S., Wang, Y., Yun, M.K., White, S.W., Parmar, K., Meibohm, B., Hartman, K.L., Wu, Z., Miller, D.D., Li, W.(2022) Cancer Lett 555: 216046-216046
- PubMed: 36596380
- DOI: https://doi.org/10.1016/j.canlet.2022.216046
- Primary Citation of Related Structures:
8DIQ - PubMed Abstract:
Extensive preclinical studies have shown that colchicine-binding site inhibitors (CBSIs) are promising drug candidates for cancer therapy. Although numerous CBSIs were generated and evaluated, but so far the FDA has not approved any of them due to undesired adverse events or insufficient efficacies. We previously reported two very potent CBSIs, the dihydroquinoxalinone compounds 5 m and 5t. In this study, we further optimized the structures of compounds 5 m and 5t and integrated them to generate a new analog, SB226. X-ray crystal structure studies and a tubulin polymerization assay confirmed that SB226 is a CBSI that could disrupt the microtubule dynamics and interfere with microtubule assembly. Biophysical measurements using surface plasmon resonance (SPR) spectroscopy verified the high binding affinity of SB226 to tubulin dimers. The in vitro studies showed that SB226 possessed sub-nanomolar anti-proliferative activities with an average IC 50 of 0.76 nM against a panel of cancer cell lines, some of which are paclitaxel-resistant, including melanoma, breast cancer and prostate cancer cells. SB226 inhibited the colony formation and migration of Taxol-resistant A375/TxR cells, and induced their G2/M phase arrest and apoptosis. Our subsequent in vivo studies confirmed that 4 mg/kg SB226 strongly inhibited the tumor growth of A375/TxR melanoma xenografts in mice and induced necrosis, anti-angiogenesis, and apoptosis in tumors. Moreover, SB226 treatment significantly inhibited spontaneous axillary lymph node, lung, and liver metastases originating from subcutaneous tumors in mice without any obvious toxicity to the animals' major organs, demonstrating the therapeutic potential of SB226 as a novel anticancer agent for cancer therapy.
- Department of Pharmaceutical Sciences, College of Pharmacy, University of Tennessee Health Science Center, Memphis, TN, 38163, United States.
Organizational Affiliation: